The purpose of the cov19 package is to integrate the differential equations of epidemiological models and fit their respective parameters with data using MCMC approach.
You need Python 3.7 or later to use cov19. You can find it at python.org. You also need setuptools, pandas, pygtc and tqdm packages, which is available from PyPI. If you have pip, just run:
pip install pandas
pip install pygtc
pip install setuptools
pip install tqdm
You can install the package via PyPI:
pip install cov19
Alternatively, you can clone this repo to your local machine using:
git clone https://github.com/emdemor/cov19
- Support for different epidemiological models
- Monte Carlo Markov Chains (MCMC) approach to fit patameters
- Data from multiple trusted and reliable sources compiled by Microsoft and accessible in www.bing.com/covid.
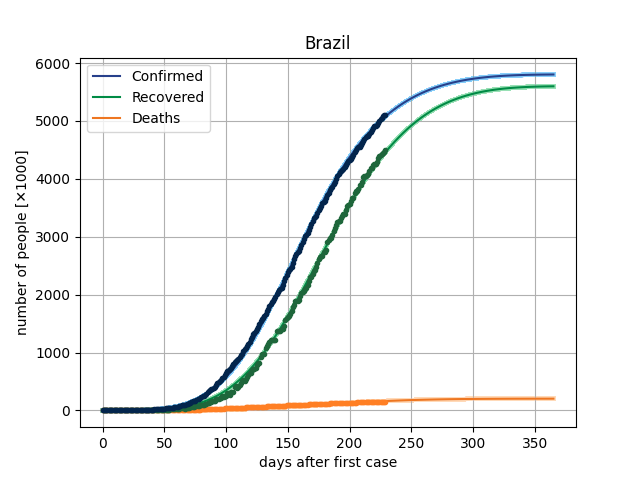
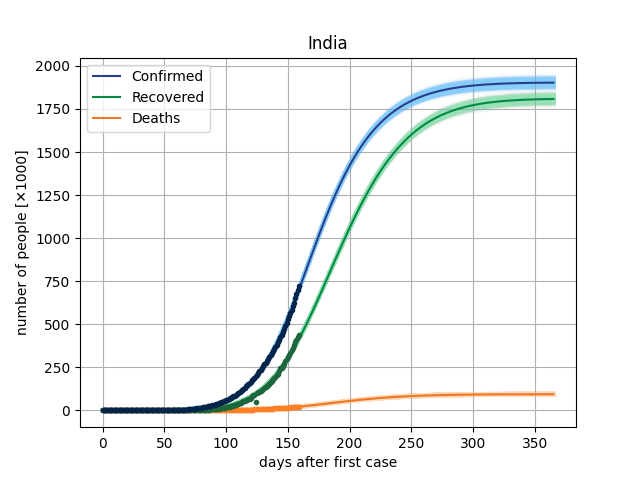
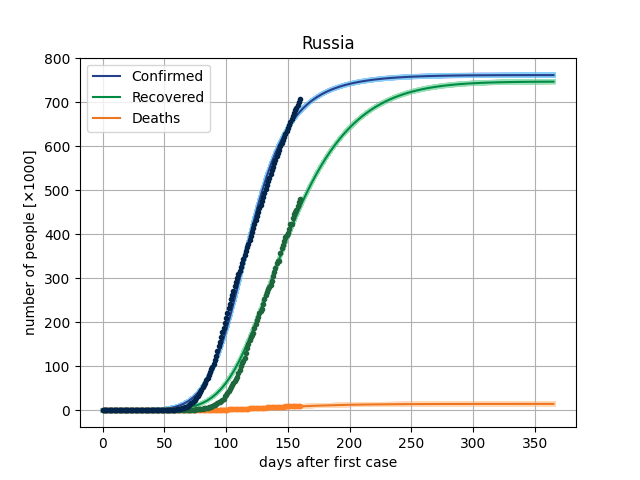
The main result in this version is to plot de curves from the model for a specific parameter vector and compare this with dataset. In covid/stat.py, functions has been implemented to generate an MCMC sample, through which it will be possible to make inferences of the parametric intervals.
Here, we are using the Microsoft Data, from the repo https://github.com/microsoft/Bing-COVID-19-Data. ]