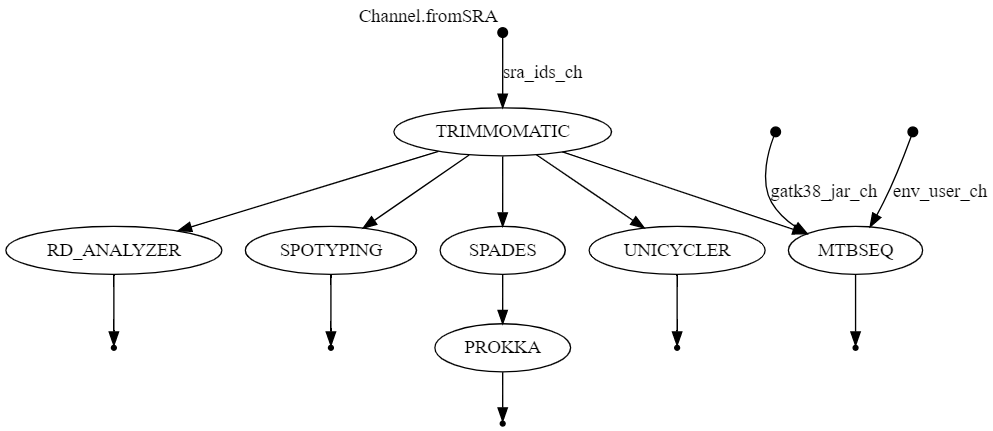
a pipeline for assemblying, anotating Mycobacterium Tuberculosis genomes and function prediction of antibiotic resistance that takes sra ids and handles the fastqs as inputs. it also integrates rd analyzer as a tool.
- Nextflow VERSION > 20.11
- Java 8
- Docker
This is the complete workflow of this pipeline.
-
Install nextflow
Please refer to Nextflow page on github for more info.
-
Run it!
nextflow run https://github.com/bioinformatics-lab/rita_bahia_analysis.git -params-file params/YOUR_PARAMS_FILE
$YOUR_PARAMS_FILE = FILE NAME, replace for your params file location, please refere to params_stardard.yml as template.
You can use diferent profiles for this pipeline, based on the computation enviroment at your disposal. Here are the Avaliable Profiles:
-
aws
-
gls
-
azureBatch
-
awsBatch
Note: Update conf/profile with your own credentials
This Pipeline can be launched on Tower, please refer to Tower launch documentation for step-by-step execution tutorial.
When launching from Tower, please update and use the params.yml file contents.
This project has the -stub-run feature, that can be used for testing propouse, it can be used on Tower with the Advanced settings on launch. You can also test it locally, using the following command:
bash data/mock_data/generate_mock_data.sh
nextflow run main.nf \
-params-file params/stub_params.yaml \
-stub-run
Problematic samples:
- ERR1035303
- ERR1035365