- About
- What To Install
- Before You Use
- Getting Ready for Your System
⚠️ File Naming Warning⚠️ - Using Snakemake
- Running on a Cluster (But Not the Snakemake Way)
- Citations
- TODOs
This repository is a Snakemake workflow for working with AMBER MD analysis.
You will need to
install snakemake
with Python 3.x for your system.
Different components of this workflow also require an installation of R.
Currently, the Residue_E_Decomp_openmp.f90 in the scripts/ directory is a
blank file for testing.
You can download the actual script from
the Cisneros Group's GitHub.
The base directory is the entry point for this workflow.
analysis: contains the output from analyses.config: configuration files. The.tsvfiles should be used to give the workflow a "map" of your directory tree.rules: contains the rules forsnakemakeand any required function definitions.scripts: the base scripts that generate system-specific scripts. This is where you would modify specific analyses or figure-rendering. They are written to take system arguments for the things specified in theconfig/config.yamlfile that would be unique for a given project.
It is recommended that you use the PyCharm (Community) IDE when editing these files, and adding the recommended plugins for Snakemake and TSV files. This will make debugging way, way easier!
Following the snakemake recommendations, you should set up a conda
environment using Python 3.6 or newer.
# Create a new environment named `snakemake` and install `snakemake` into it
$ conda -n snakemake snakemake
# Activate the environment
$ conda activate snakemake
# Install the packages needed for plotting scripts
# `pandas` and `numpy` should already be `snakemake` dependencies
$ conda install matplotlib
$ conda install prody
$ conda install statsmodels
$ conda install gnuplot
# Install the things necessary for R (required for `eda_avg` rule)
$ conda install r-base r-essentials
$ conda install r-abindYou can build this environment simply by running:
$ conda env create -f env/environment.ymlIf you are going to install this today, you will want to use the GitHub
version of Prody due to an error when parsing NMD file in the 2.0 release.
Instead of conda install prody, instead do:
$ conda activate snakemake
$ conda install git pip
$ pip install git+https://github.com/prody/ProDy.git
The gnuplot install is not required if you already have a gnuplot
installation on the system you will use.
If you already have an R installation, you don't have to go through the conda
R installation steps.
In that case, if R is stored a module, source the module before running
the workflow with snakemake.
If you would rather have everything contained in conda, make sure you
comment out any ~/.Renviron file you may have, as it will point to the wrong
library path, and those libraries are likely incompatible with the
conda-installed version.
The scripts/ directory contains the scripts for the analysis.
If you have specific analyses or plotting needs, you will want to modify
these scripts.
For example, if you want to plot distances for a specific atom pair,
you will want to add that into the write_analy_traj function of
write-cpptraj.py.
Similarly, if you know you need specific axes for plots, you will want to
add those to the relevant scripts.
If you want to set up variables as part of a .tsv, then that process will
require a bit more work, including:
- Adding the code to read the new
.tsvstarting with aPathcolumn that matchessystems.tsv - Storing the newly read
.tsvas a dictionary - Get snakemake to parse the dictionary when iterating through the systems
- Adding system arguments to the Python scripts
- Adding those system arguments into the appropriate
rules/*.smkfile
The config directory contains 3 files:
-
config.yaml: contains options and descriptions for configuring the workflow -
EDAvalues.tsv: a tab separated file with 5 columns of information necessary to set up the files for Energy Decomposition Analysis (EDA). Keep the header! These columns include:Path: The path within the analysis tree where the files should be saved. A typical path should look likeSystem/replicate. This block must match thePathspecified insystems.tsv.NRES: The number of non-solvent residues that you want to look at for the EDA. (Ex: 455)NATOM: The total number of atoms in the trajectory. (Ex: 51348)NPROTAT: The number of atoms in the non-solvent portion of residues selected inNRES. (Ex: 5880)TOTRES: The number of total residues in the trajectory. (Ex: 20348)
NRESandNATOMcan be identified from the.prmtopfile, but typically the system will need to be converted to a PDB to determineNRESandTOTRES. -
systems.tsv: a tab separated file with 5 columns of information necessary to set up the file paths for various analyses. This is effectively a roadmap for the directory tree. Keep the header!-
Path: The path within the analysis tree where the files should be saved. A typical path should look likeSystem/Replicate. This block must match thePathspecified inEDAvalues.tsv.(Ex:
WT_r1) -
System: The system that the analysis is being performed on. This should match the first part ofPath(prior to the slash). An example of different systems would beWT,MUTA, andMUTB.(Ex:
WT) -
Replicate: The replicate. For the R-based EDA rules, particularly, you need at least 3 replicates. This should match the second part ofPath(after the slash).(Ex:
r1) -
Parm_Path: The full system path to the trajectory files for a particularSystem/Replicate. Theprmtop,inpcrd, andncormdcrdfiles should be in the same directory.(Ex:
/home/$USER/project/system/replicate) -
Sys_Tag: The "tag" for files written in a shared directory that distinguishes one system type from another. Typically, this will look like a combination of thePROJ_IDspecified in theconfig/config.yamland theSystem.(Ex:
ProteinID_WT)
-
You MUST have consistent file naming for the prmtop, inpcrd, and trajectory
files!
THIS IS CRUCIAL TO THE WORKFLOW!!!!!
The Sys_Tag is specified in the config/systems.tsv file.
You can modify the scripts a little bit, but it's set up for stuff like this:
-
Prmtop:
{Sys_Tag}.prmtop- Ex reps 1: crambin-WT-wat.prmtop, crambin-H39C-wat.prmtop
- Ex reps 2: crambin_WT.prmtop, crambin_H39C.prmtop
-
Inpcrd:
{Sys_Tag}.inpcrd- Ex reps 1: crambin-WT-wat.inpcrd, crambin-H39C-wat.inpcrd
- Ex reps 2: crambin_WT.inpcrd, crambin_H39C.inpcrd
-
Traj (mdcrd/nc):
{Sys_Tag}{fs}md{num}.{f_ext}- Ex reps 1: crambin-WT-wat-md50.mdcrd, crambin-H39C-wat-md50.mdcrd
- Ex reps 2: crambin_WT_md50.nc, crambin_H39C_md50.nc
| Variable | Explanation |
|---|---|
Sys_Tag |
The project identifier for a system/replicate. |
fs |
A file separator. Common examples are -, _, and . |
system |
What system the files are for (like wild type or a specific mutant). |
num |
The number of the trajectory file, since we write in short chunks. |
f_ext |
The file extension type. You might save using nc or mdcrd. |
Basically, don't interchange between the examples. If you did, you'll want to rename all your files SAFELY. Do NOT think "oh this loop is safe" without testing it AWAY from your data first!!! You may think it'll work fine, but that's a really easy way to overwrite or delete your data in 10 seconds.
The rename command (which doesn't exist on all operating systems...) can be
useful for doing this.
It takes the current naming you want to switch, the thing you want to switch
it to, and the files it should be applied to as arguments.
$ ls
thing-name-2020-want.inpcrd thing-name-2020-want_md3.nc
thing-name-2020-want_md1.nc thing-name-2020-want.prmtop
thing-name-2020-want_md2.nc
$ rename thing-name-2020-want proteinID-WT-wat thing-name-2020-want*
$ ls
proteinID-WT-wat.inpcrd proteinID-WT-wat_md2.nc proteinID-WT-wat.prmtop
proteinID-WT-wat_md1.nc proteinID-WT-wat_md3.nc
$ rename _md -md proteinID-WT-wat_*
$ ls
proteinID-WT-wat.inpcrd proteinID-WT-wat-md2.nc proteinID-WT-wat.prmtop
proteinID-WT-wat-md1.nc proteinID-WT-wat-md3.ncIf you used prod or didn't use a word to specify production in your
trajectory files, you may opt to remove the md specifier in the scripts and
files, but it can be a pain.
Using rename is certainly easier.
You can use snakemake -np as a dry-run.
It will verify that all files are present and show commands to be executed.
If files are missing in the expected paths, it will print a warning.
Each rule can be used alone with something like:
snakemake <rule> --cores 1
There's a whole host of different options in the snakemake documentation.
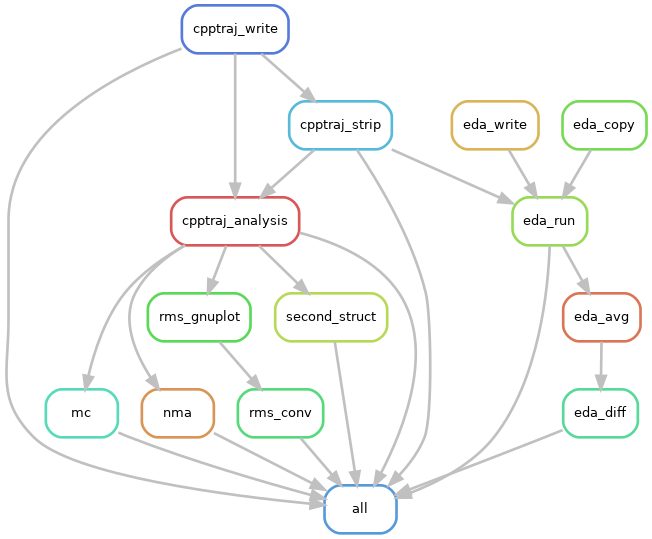
You can create a PDF of how Snakemake thinks your workflow links together:
snakemake --forceall --dag | dot -Tpdf > dag.pdf
You can also make a PNG by changing the file type.
snakemake --forceall --dag | dot -Tpng > dag.png
This is a great way to check for errors!
You can test deleting output from snakemake with:
snakemake cpptraj_write --delete-all-output --dry-run
And actually do it with:
snakemake cpptraj_write --delete-all-output --cores 1
This particular workflow has a clean rule, which will remove the previous
analyses.
Be careful, though, as this will remove any generated input scripts or
trajectory files.
You can rewrite the rule for yourself in rules/common.smk.
snakemake clean --cores 1
Part of what this workflow does is build the specific queue submission scripts
for each job, since each job type has different needs.
While snakemake can run through a cluster system, this approach seemed ideal
when dealing with specific workflow rules, especially those pertaining to EDA.
However, because of this atypical approach to snakemake, we want to set the
output-wait flag so that it knows that it might take a while for certain
jobs to run.
This may backfire!
If you don't see any other jobs in the queue from the snakemake job for a
few minutes, you may want to end the job and investigate.
Similarly, because interactive jobs are dependent on the VPN, we can submit
the overall snakemake job through the queuing system.
#!/bin/bash
#PBS -q my_cpu_alloc
#PBS -l nodes=1:ppn=2,mem=8GB
#PBS -j oe
#PBS -r n
#PBS -o err.error
#PBS -N proteinID_smk
cd $PBS_O_WORKDIR
## Use this to evaluate the conda commands
# eval "$(conda shell.bash hook)"
source ~/.bash_profile
## Set-up conda environment
# conda create -n snakemake snakemake
## Activate the conda environment
conda activate snakemake
## Run snakemake
## Output wait time is in seconds
## Wait 2 hours for new files: 7200
## Wait 24 hours for new files: 86400
snakemake --cores 2 --output-wait 7200- Cite
snakemakefor their powerhouse software - Cite
pandasfor their beautiful TSV reading functions - Cite
numpyfor all things math - Cite
matplotlibfor the figures it helped make - Cite
statsmodelsfor their hand in the matrix correlation figures - Cite
prodyfor doing the normal mode analysis
- Cite
Ritself for existing in the world - Cite
tidyversefor changing the game of data table processing - Cite
data.tablefor reading EDA data with ease - Cite
abindfor helping to process the EDA data
- The
Residue_E_Decomp_openmp.f90program (an empty file here to check the workflow) can be downloaded from the Cisneros Group's GitHub. The citation information is shared there.
- Write some scripts for renaming files to the syntax required for this workflow
- Add more analysis and test on a real system
- Test on a cluster
- Create (and test!) the environment files
- Add SLURM and LSF examples to scripts for bash