Citation
If you use the proposed model in your research or wish to refer to the results published, please use the following BibTeX entry.
@article{altinsoy2021improved,
title={An improved denoising of G-banding chromosome images using cascaded CNN and binary classification network},
author={Altinsoy, Emrecan and Yang, Jie and Tu, Enmei},
journal={The Visual Computer},
pages={1--14},
year={2021},
publisher={Springer}
}
Setup Environment
- Cuda 10.2
- Cudnn v8.1.0.77
conda create -n <your_env_name> python=3.8activate <your_env_name>conda install pytorch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0 cudatoolkit=10.2 -c pytorchconda install -c anaconda pillow scikit-image scikit-learn imageio pandas seabornconda install -c conda-forge pytorch-model-summary tensorflow==1.15pip install ptflopsTo use cloud TPU run the code below to install torch_xla before running the tpu_train.py and tpu_inference.py files
!pip install cloud-tpu-client==0.10 https://storage.googleapis.com/tpu-pytorch/wheels/torch_xla-1.8-cp37-cp37m-linux_x86_64.whlDataset Preparation
Before running classification_train.py, classification data needs to be created. To create the classification dataset, you need to run classification_data_prep.py file. Run the following command to see the details of the arguments.
python3 ./src/classification_data_prep.py -h
python3 ./src/classification_data_prep.py --model=preactivation_resunet --weight-path=preactivation_resunet-20211012T1603 --weight-num=40 --images='datasets/raw_chromosome_data/'
Training
usage: ./src/segmentation_train.py [-h] [--model MODEL] [--pretrained PRETRAINED]
[--batch-size BATCH_SIZE] [--epochs EPOCHS]
[--lr LR] [--device DEVICE] [--workers WORKERS]
[--weights WEIGHTS] [--test TEST] [--logs LOGS]
[--images IMAGES] [--image-size IMAGE_SIZE]
[--init-features INIT_FEATURES]
Semantic segmentation of G-banding chromosome Images
optional arguments:
-h, --help show this help message and exit
--model MODEL choose model to train
--pretrained PRETRAINE is the backbone pretrained or not
--batch-size BATCH_SIZ input batch size for training (default: 2)
--epochs EPOCHS number of epochs to train (default: 40)
--lr LR initial learning rate (default: 0.0001)
--device DEVICE device for training (default: cuda:0)
--workers WORKERS number of workers for data loading (default: 0)
--weights WEIGHTS folder to save weights
--test TEST folder to save weights
--logs LOGS folder to save logs
--images IMAGES dataset folder directory
--image-size IMAGE_SIZE target input image size (default: 480x640)
--init-features INIT_FEATURES init features for unet, resunet, preact-resunet
available models:
unet, resunet, preactivation_resunet, cenet, segnet, nested_unet,
attention_unet, fcn_resnet101, deeplabv3_resnet101, pspnet
usage: classification_train.py [-h] [--batch-size BATCH_SIZE]
[--epochs EPOCHS] [--lr LR]
[--device DEVICE] [--workers WORKERS]
[--weights WEIGHTS] [--logs LOGS]
[--train-data TRAIN_DATA]
[--validation-data VALIDATION_DATA]
Classification of chromosome and non-chromosome objects
optional arguments:
-h, --help show this help message and exit
--batch-size BATCH_SIZE input batch size for training (default: 20)
--epochs EPOCHS number of epochs to train (default: 100)
--lr LR initial learning rate (default: 0.0001)
--device DEVICE device for training (default: cuda:0)
--workers WORKERS number of workers for data loading (default: 0)
--weights WEIGHTS folder to save weights
--logs LOGS folder to save logs
--train-data TRAIN_DATA directory of training data
--validation-data VALIDATION_DATA directory of validation data
python3 ./src/segmentation_train.py --images='datasets/raw_chromosome_data/' --model='unet' --epochs=50 --batch-size=4
python3 ./src/classification_train.py --train-data=datasets/binary_classification_data/train_data.csv --validation-data=datasets/binary_classification_data/valid_data.csv --epochs=100 --batch-size=40
Results
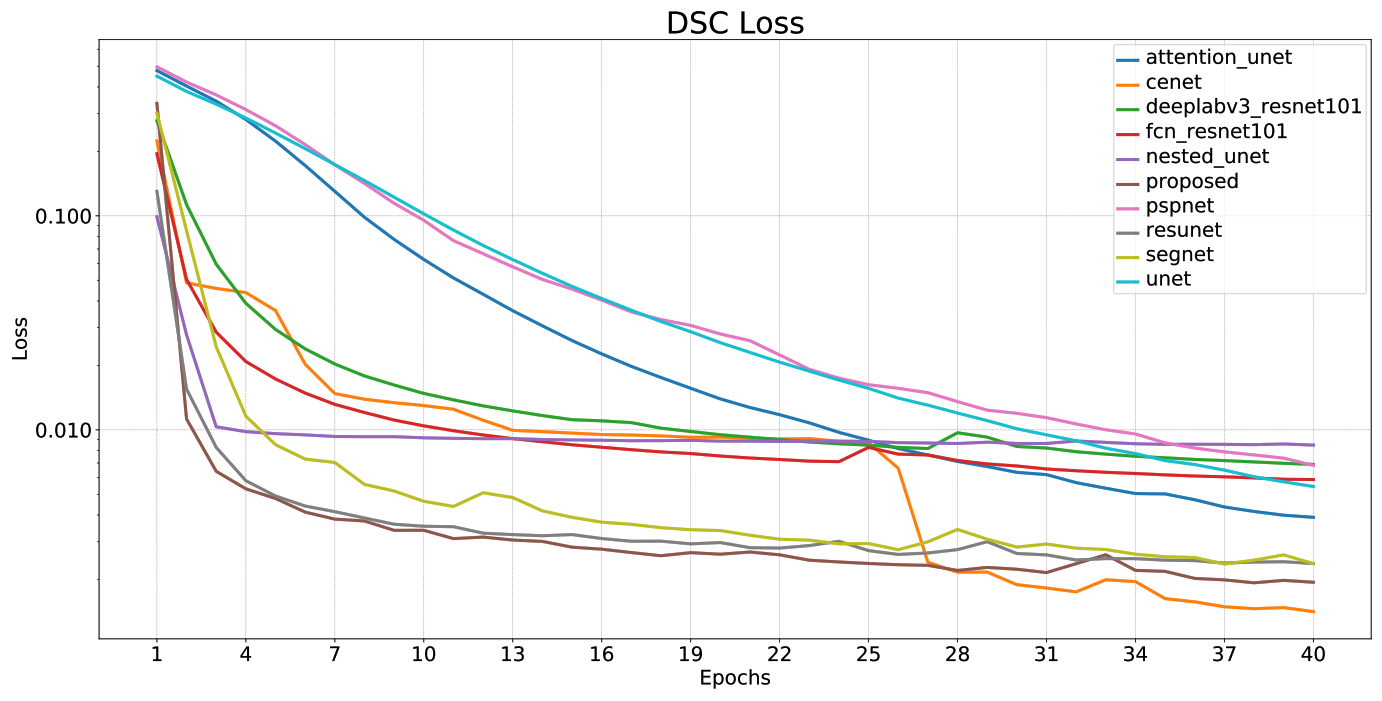 |
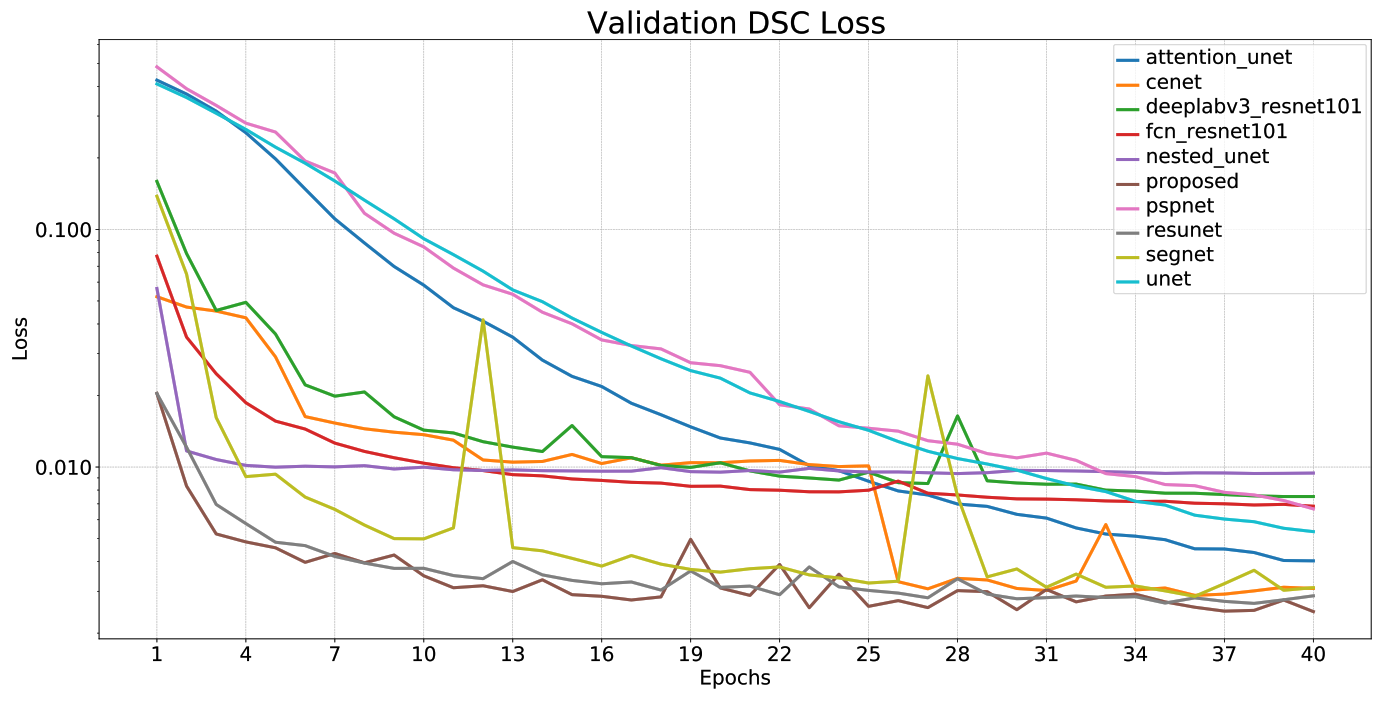 |
| Model | Params | MACs | Dice (%) | Se (%) | Sp (%) | Pre (%) | Acc (%) |
|---|---|---|---|---|---|---|---|
| U-Net | 7.76 M | 64.24 GMac | 99.4998 | 99.5379 | 99.7306 | 99.4616 | 99.6664 |
| Residual U-Net | 8.11 M | 67.39 GMac | 99.7657 | 99.7649 | 99.8833 | 99.7666 | 99.8438 |
| U-net++ | 9.05 M | 158.91 GMac | 98.9878 | 98.1763 | 99.9079 | 99.8128 | 99.3307 |
| Attention U-net | 34.88 M | 312.04 GMac | 99.6272 | 99.6367 | 99.8088 | 99.6177 | 99.7515 |
| CE-Net | 29.0 M | 41.89 GMac | 99.7414 | 99.7416 | 99.8706 | 99.7412 | 99.8276 |
| SegNet | 29.44 M | 188.33 GMac | 99.7415 | 99.7511 | 99.8659 | 99.7319 | 99.8277 |
| FCN8s (Resnet-101) | 51.94 M | 253.85 GMac | 99.3271 | 99.3324 | 99.6609 | 99.3218 | 99.5514 |
| DeepLab v3 (Resnet-101) | 58.63 M | 283.44 GMac | 99.2612 | 99.2848 | 99.6186 | 99.2377 | 99.5074 |
| PSPNet (Resnet-101) | 72.31 M | 327.25 GMac | 99.3541 | 99.3847 | 99.6615 | 99.3234 | 99.5693 |
| Proposed CNN | 8.11 M | 67.44 GMac | 99.7836 | 99.787 | 99.8901 | 99.7802 | 99.8557 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Model | Dice (%) | Se (%) | Sp (%) | Pre (%) | Acc (%) |
|---|---|---|---|---|---|
| Local Adaptive Thresholding | 76.1106 | 71.8094 | 99.3036 | 84.3697 | 97.5273 |
| Histogram Analysis | 81.3935 | 79.8560 | 99.3562 | 85.4999 | 98.1877 |
| U-Net (t=18) | 98.1012 | 98.1355 | 99.9144 | 98.072 | 99.8397 |
| Residual U-Net (t=187) | 97.9031 | 97.8826 | 99.908 | 97.9306 | 99.8227 |
| U-Net++ (t=171) | 98.084 | 98.1604 | 99.9115 | 98.0139 | 99.8378 |
| Attention U-Net (t=252) | 97.8263 | 97.6727 | 99.9116 | 97.9905 | 99.8163 |
| CE-Net (t=190) | 97.677 | 97.6342 | 99.899 | 97.7274 | 99.8039 |
| SegNet (t=173) | 97.6706 | 97.6965 | 99.8959 | 97.6528 | 99.8033 |
| FCN8s (t=109) | 93.3623 | 92.2031 | 99.7601 | 94.5663 | 99.4357 |
| DeepLab v3 (t=124) | 93.1646 | 91.8936 | 99.757 | 94.4835 | 99.4186 |
| PSPNet (t=180) | 95.2566 | 95.1396 | 99.7966 | 95.386 | 99.6006 |
| Proposed CNN (t=131) | 98.0006 | 97.9824 | 99.9121 | 98.0316 | 99.832 |
| Proposed CNN + BCN | 98.735 | 98.6783 | 99.9467 | 98.7918 | 99.8931 |























































