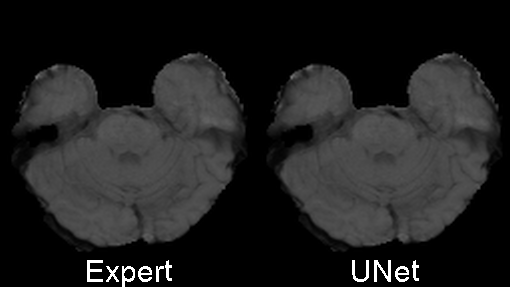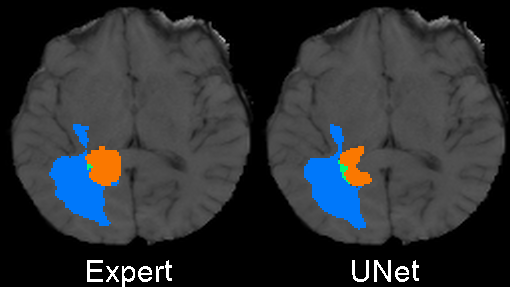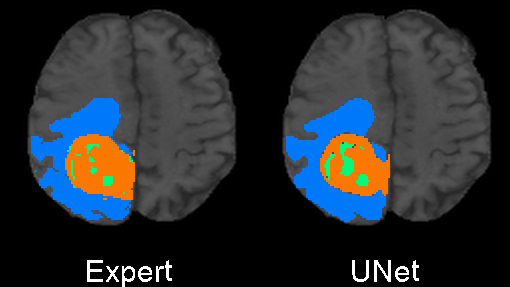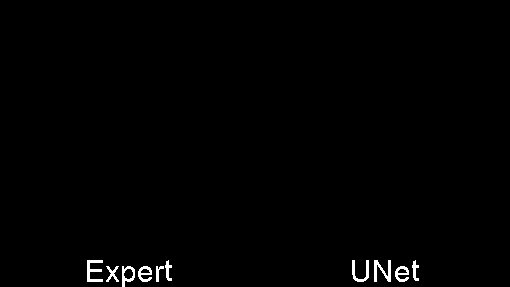
The code was written to be trained using the BRATS 2020 data set for brain tumors, but it can be easily modified to be used in other 3D applications.
- Download the BraTS 2020 data after registering by following the steps outlined on the BraTS 2020 competition page.
Place the unzipped training and validation data folders named "MICCAI_BraTS2020_TrainingData"
and "MICCAI_BraTS2020_ValidationData" in the
brats/datafolder. - Install Python 3 and dependencies:
- nibabel
- keras
- pytables
- nilearn
- SimpleITK
- keras-contrib
-
cdinto the 3DUnetCNN repository. -
Add the repository directory to the
PYTONPATHsystem variable:
$ export PYTHONPATH=${PWD}:$PYTHONPATH
-
cdinto thebratsfolder. -
Run the training:
$ python train.py
- Now that the model is trained, predict the BraTS validation data:
$ python predict.py
The predicted segmentations will be in the "BraTS2020_Validation_predictions".
If you run out of memory during training: try setting
config['patch_shape`] = (64, 64, 64) for starters.
Also, read the "Configuration" notes at the bottom of this page.
In the training above, part of the data was held out for validation purposes. To write the predicted label maps to file:
$ python predict.py
The predictions will be written in the prediction folder along with the input data and ground truth labels for
comparison.
Changing the configuration dictionary in the train.py scripts, makes it easy to test out different model and
training configurations.
I would recommend trying it out then modifying the parameters until you have satisfactory
results.
If you are running out of memory, try training using (64, 64, 64) shaped patches.
Reducing the "batch_size" and "validation_batch_size" parameters will also reduce the amount of memory required for
training as smaller batch sizes feed smaller chunks of data to the CNN.
If the batch size is reduced down to 1 and it still you are still running
out of memory, you could also try changing the patch size to (32, 32, 32).
Keep in mind, though, that a smaller patch sizes may not perform as well as larger patch sizes.
If you want to train a 3D UNet on a different set of data, you can copy either the train.py script and modify it to read in your data rather than the preprocessed BRATS data that they are currently setup to train on.
The following Keras model were trained on the BRATS 2020 data:
| Dice Enhancing Tumor | Dice Whole Tumor | Dice Tumor Core |
|---|---|---|
| 0.65446 | 0.86171 | 0.75757 |