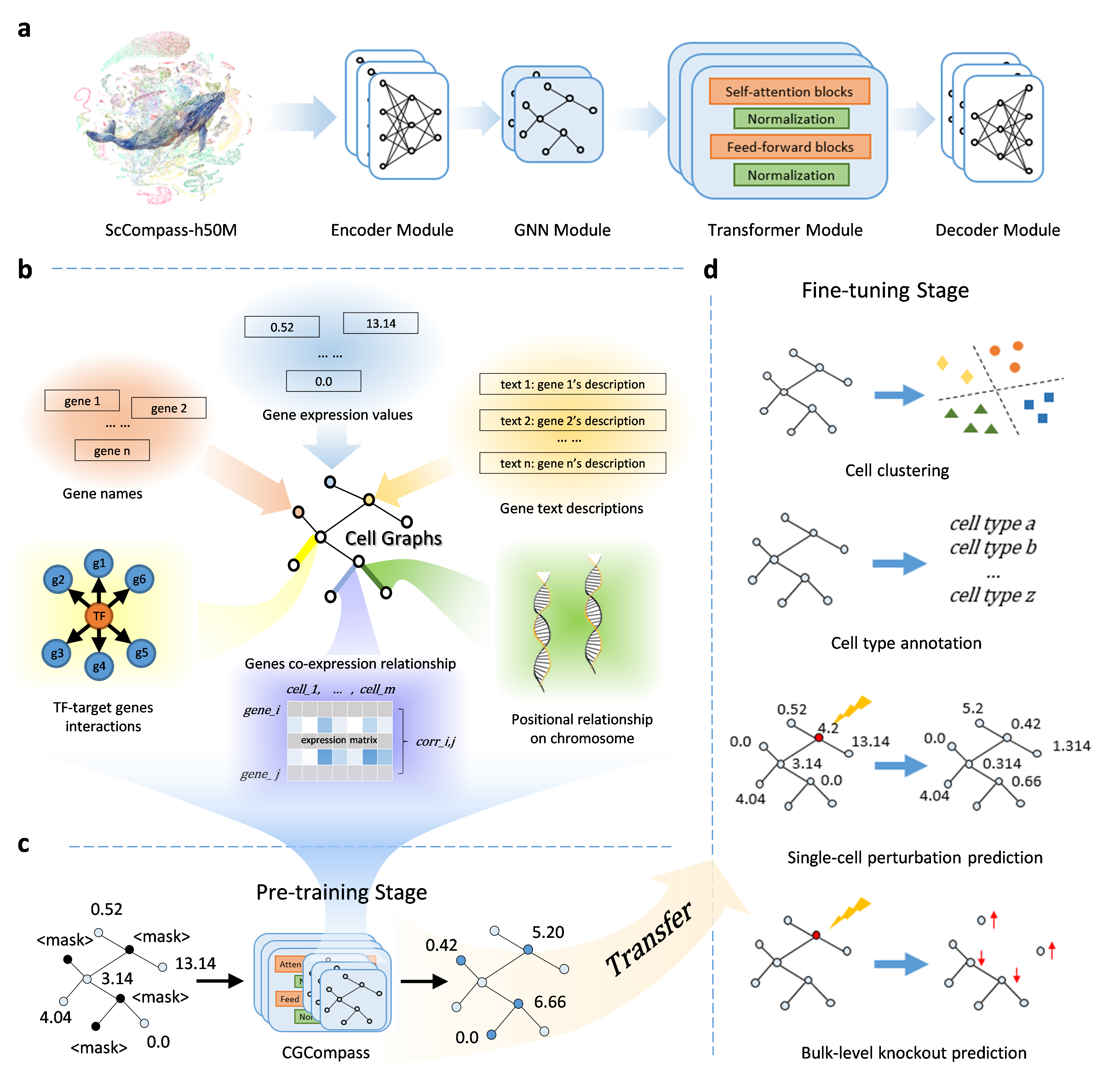
This is the open source code repository for the paper "Cell-GraphCompass: Modeling Single Cells with Graph Structure Foundation Model." Cell-GraphCompass (CGCompass) is a graph-structured single-cell transcriptome foundational model. It uses six biological features to construct cell graphs and is pre-trained on fifty million human single-cell sequencing data.
git clone https://github.com/epang-ucas/Cell-Graph-Compass.git
cd Cell-Graph-Compass
conda create -n CGC python=3.9.16 R
conda activate CGC
pip install torch==1.13.0
pip install -r requirements.txtWe use the Uni-Core framework to organize the code. You can download it from here and install it as follows:
cd Uni-Core-0.0.1
python setup.py install
cd ../exps: Contains shell scripts to start model training and testing./src: Contains the source code./scData: Contains data required by model training or testing.
Due to the large size of the model's weight parameters and database files, we stored them on Google Drive. Users should download them first and then place them in the /scData directory.
The model's training and testing can be initiated by shell scripts. Below are example scripts for three downstream tasks.
exps/cellClus.sh: Cell clustering and batch correction.exps/cellAnno.sh: Cell type annotation.exps/pert_norman: Single-cell gene perturbation prediction.
The scripts include the hyperparameters used in the experimental results presented in our paper, and users can adjust them as needed.
As an example, for cell clustering on the PCortex dataset, follow these steps:
First, preprocess the original dataset as described in scData/readme.md.
We also provide a processed example database at ./scData/example_datasets/cellclus_PCortex.
Then, start the training and testing script:
bash ./exps/cellClus.shPlease cite us as follows:
@article {Fang2024.06.04.597354,
author = {Fang, Chen and Hu, Zhilong and Chang, Shaole and Long, Qingqing and Cui, Wentao and Liu, Wenhao and Li, Cong and Liu, Yana and Wang, Pengfei and Meng, Zhen and Pan, Jia and Zhou, Yuanchun and Feng, Guihai and Chen, Linghui and Li, Xin},
title = {Cell-Graph Compass: Modeling Single Cells with Graph Structure Foundation Model},
elocation-id = {2024.06.04.597354},
year = {2024},
doi = {10.1101/2024.06.04.597354},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/06/06/2024.06.04.597354},
eprint = {https://www.biorxiv.org/content/early/2024/06/06/2024.06.04.597354.full.pdf},
journal = {bioRxiv}
}