The goal of this library is to interact with a Miro board that has
been set up to represent a Bayesian network. We assumed that in
Miro, sticky notes represent the network’s nodes, and
connectors represent arcs suggesting a causal/influence link. Sticky
notes should have text describing the node, and a single tag,
indicating the name of the corresponding variable in the database.
With this library, you will be able to gather the attributes to lay out
the DAG (Directed Acyclic Graph) of the network, which has been
collaboratively portrayed in Miro. With all the basic data brought
into R, you can formally process a DAG. This includes the ability to
further process and train the model in R, and even in other platforms
like bnlearn in R, or the various options available in Python or
in dedicated applications like Netica. There are functions in this
library to do so.
You can install the development version of miro2bayes like so:
library(devtools)
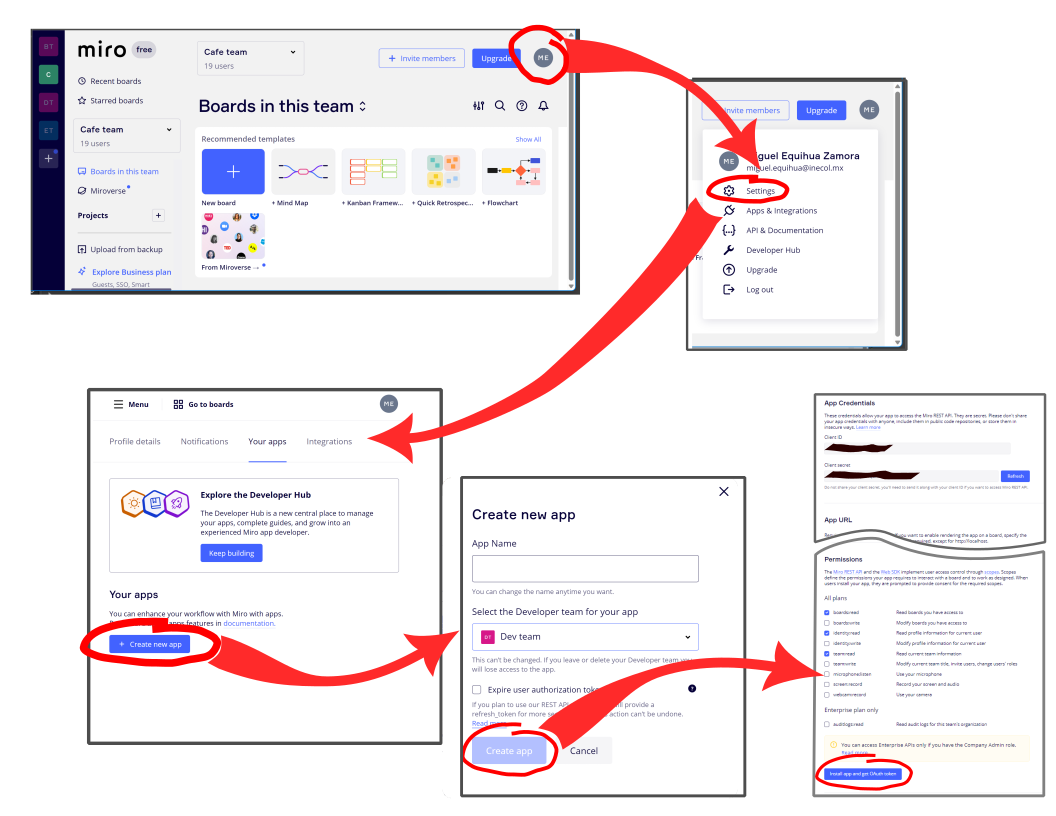
install_github("equihuam/miro2bayesNet")To interact with Miro you need your credentials, which can be obtained following this Quickstart directions for your first REST API call. A summary of the steps involved is shown in the Figure below.
The miro2bayes library uses keyring library to keep your credentials
safe; so, you will need to register your credentials in your local
machine with the function
key_set(service="identifier-you-like", username = "user-name-you-like")
as described here. Both
service and username are required, respectively, as servMiro and
user parameters, to get access to Miro with the function getMiro
as explained below.
This is a basic example which shows you how to get Bayesian network data from Miro:
miro_data <- getMiro(servMiro = "miro", user = "your-miro-token",
board = tablero_tr)
miro_dataIf you need to find the board id of interest, you could use the
function miroBoards. This function queries Miro with the
credentials provided and returns a tibble with the data describing the
boards available to that user.
miro_boards <- miroBoards(servMiro = "miro", user = "your-miro-token")
miro_boards[, c("name", "id")]
# You could select what yyyou whant this way
board_item <- miro_boards %>%
filter(str_detect(name, {"target frase"})) %>%
select(id, name)A typical cycle of use would be as illustrated in the Figure. This way you shall have a collection of implied conditional independence relations to discuss the DAG structure. By critical analysis of the DAG you should go back to Miro to solve inconsistencies or further develop the intended causal proposition.
miro_data <- getMiro(servMiro = "miro", user = "your-miro-token",
board = tablero_tr)
names(miro_data$dag)A quick check of the network recovered from Miro can be produced
with the function miroValidation. The check shows whether the network
is indeed a DAG (no cycles present!) and a few numbers describing what
was found in Miro: number of nodes, identified variable names, the
status of links, and so for.
miroValidation(miro_data))Once a satisfactory Bayesian network has been produced in Miro, the function miro2DNE is used to produce a .DNE file that can be used in Netica and other dedicated Bayesian network software for training, analysis and prediction.
data_DNE <- miro2DNE(miro_data)Another option is to feed Miro data into bnlearn with the function
miro2bnlearn. Which is done as follows.
netMiro_bnl <- miro2bnlearn(miro_data)One interesting option you have, once a DAG is available in R is the
identification of implied conditional independence patterns. This is
done by dagitty, but you can subset them for convenience with
miro2bayes. All you have to do is provide the name of one variable to
select all implied conditional independence expressions that relate to
that variable. The result is displayed as a web-page, to improve
formatting and hopefully, usability.
cond_indepOnvar(datos_miro, {variable})