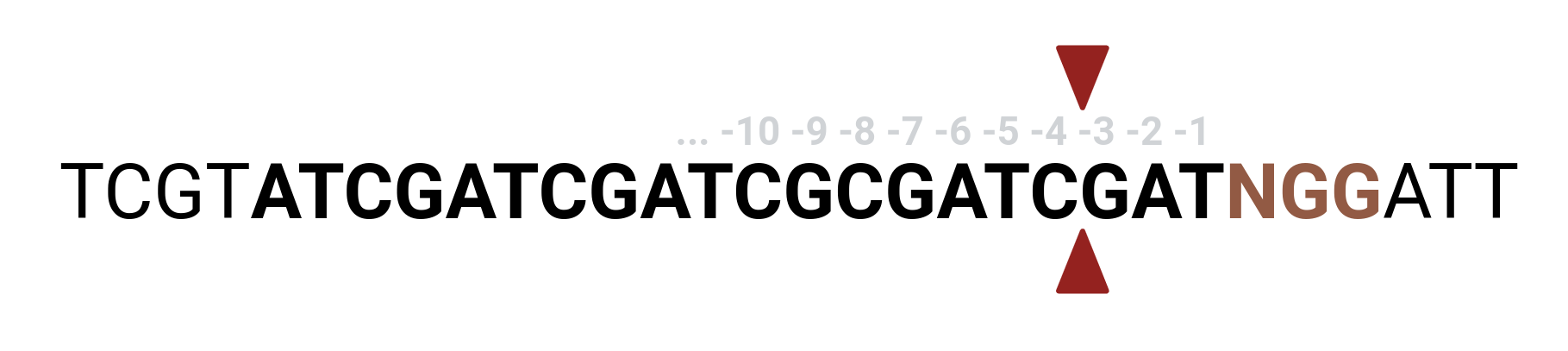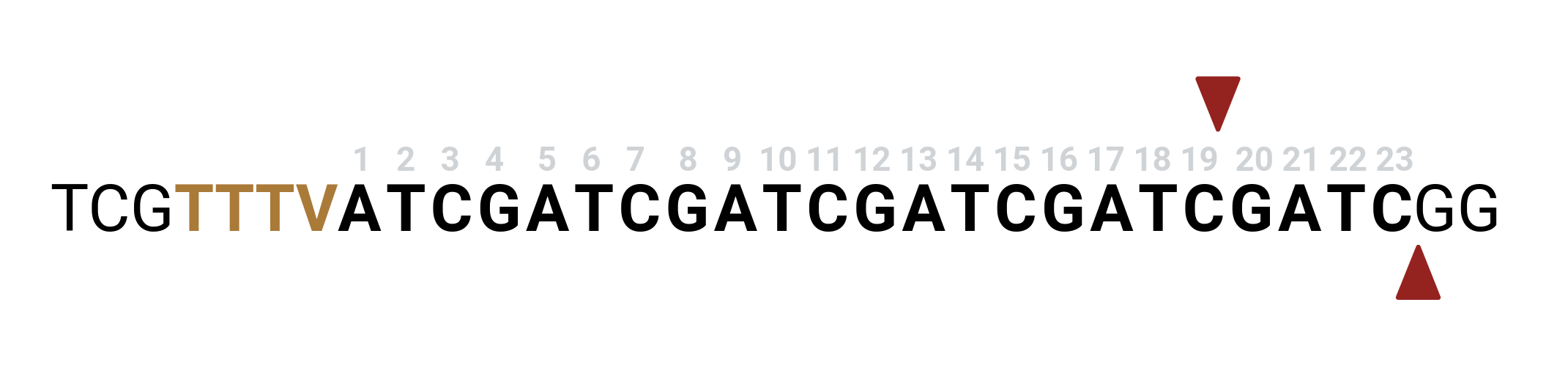CRISPRware is a comprehensive toolkit designed to preprocess NGS data and identify, score, and rank guide RNAs (gRNAs) for CRISPR experiments. It supports RNASeq, RiboSeq, ATACSeq, DNASESeq, ChIPSeq, and other genomic preprocessing techniques.
With conda installed perform the following commands.
git clone https://github.com/ericmalekos/crisprware crisprware
cd crisprware
conda env create -f environment.yml
conda activate crisprware
chmod +x setup.py
./setup.py install
- FASTA File
- BED file: A BED file can be provided to specify regions of interest within the genome. This file can help to limit the search space for gRNA identification.
- GTF/GFF file: A GTF or GFF file can be used to provide gene annotations. This information can be used to filter gRNAs based on specific genomic features such as exons or coding sequences .
We demonstrate usage with ce11 chromosome III fasta and NCBI GTF, included in the tests/test_data/ce11 directory:
Note the example off-target index is limited to chrIII, not the full ce11 genome
index_genome -f tests/test_data/ce11/chrIII_sequence.fasta
Default settings generate NGG protospacer guides
generate_guides -f tests/test_data/ce11/chrIII_sequence.fasta \
-k tests/test_data/ce11/chrIII_ce11.ncbiRefSeq.gtf \
--feature CDS
Scoring will take ~5 minutes and uses 8 threads by default.
Change this with --threads . --tracr is either Chen2013 or Hsu2013, see RuleSet3 scoring for details
score_guides -b sgRNAs/sgRNAs.bed -i GscanIndex/GscanIndex --tracr Chen2013
Ranking is done based on scoring columns
-c is matched with -m order so this filters out RS3_score_Chen2013 < 0, specificity_gscan_index < 0.2
-p 5 65, -f CDS filters out gRNAs outside of the 5th-65th percentile of the CDS
--output_all outputs TSV and histograms for each stage of filtering in addition to the final output.
rank_guides \
-k ScoredSgRNAs/ScoredSgRNAs.tsv \
-t tests/test_data/ce11/ce11.ncbiRefSeq.gtf \
-f CDS \
-c RS3_score_Chen2013 specificity_gscan_index \
-m 0 0.2 \
-p 5 65 \
-r RS3_score_Chen2013 \
--output_all
Memory requirements may be substantial in both the index_genome and score_guides steps. Guidescan2 authors provide compiled indices for some model species in the download section of their website which can be downloaded directly to avoid use of index_genome.
For score_guides we provide a parameter --chunk_size <n> which can be used to decrease memory usage by processing <n> guides at a time instead of all at once. Default setting is 100000. Increasing this number will speed up processing time and memory requirements, decreasing will slow down processing time and decrease memory requirements.
CRISPRware offers a series of modules to preprocess NGS data and determine suitable gRNAs for CRISPR applications.
The module preprocess_annotation takes processed RNASeq TPMs from Kallisto, Salmon, FLAIR, or Mandalorian from one or more samples along with the GTF/GFF gene annotation. All processed samples should be from the same quantification tool, don't mix Salmon and Kallisto files. If multiple samples are passed, max, min, median, and mean TPM values for each transcript are determined, and the user can supply minimum cut-offs for any combination of these to filter out lowly expressed isoforms. All detected isoforms (TPM > 0) are kept by default. The user can also set an integer flag --top-n <n> which will filter out all but the most highly expressed isoform for each gene. So, --top-n 1 will retain only the gene model of the most highly expressed isoform - according to median_tpm if multiple RNA seq files are passed.
There are also --tss_window and --tes_window options, which produce BED for dCas target choices. User can use these GTFs/BEDs in the generate_guides step and the rank_guides step.
preprocess_annotation -g test_data/chr19_ucsc_mm39.ncbiRefSeq.gtf \
-t quant1.sf quant2.sf quant3.sf \
--type infer \
--median 5 \
--top_n 10 \
--top_n_column median \
--model consensus metagene shortest longest \
--tss_window 300 300
--tes_window 300 300
IMPORTANT: ensure the GTF and the TPM files are have the same transcript IDs
A number of tools exists for calling translated ORFs from RiboSeq. In order to find gRNAs against these putative coding regions we can convert output from these programs into a GTF with annotated coding sequence (CDS) entries and run the pipeline normally.
For ORFs called with RiboTISH set these options in the ribotish predict command: --inframecount, --blocks, --aaseq and provide the same GTF that was passed to ribotish.
For other RiboSeq ORF callers raise a github issue and I will address it.
Full filtering options:
./scripts/gtf_from_ribotish.py -h
options:
-h, --help show this help message and exit
-r RIBOTISH, --ribotish RIBOTISH
Path to the Ribotish predict TSV file
-i INPUT_GTF, --input_gtf INPUT_GTF
Path to the corresponding GTF file
-o OUTPUT_GTF, --output_gtf OUTPUT_GTF
Path to output the new GTF file
--min_aalen MIN_AALEN
Minimum amino acid length
--min_inframecount MIN_INFRAMECOUNT
Minimum in-frame count
--max_tisqvalue MAX_TISQVALUE
Maximum TIS Q-value
--max_frameqvalue MAX_FRAMEQVALUE
Maximum Frame Q-value
--max_fisherqvalue MAX_FISHERQVALUE
Maximum Fisher Q-value
--select_based_on {AALen,InFrameCount,TISQvalue,FrameQvalue,FisherQvalue}
Column to select the best row for each Tid, TisType pair
--genetype GENETYPE GeneType to filter, must match a column entry
--tistype TISTYPE TisType to filter, must match a column entry
Default generate_guides settings are equivalent to
generate_guides \
-f <fasta> \
--pam [-p] NGG
--sgRNA_length [-l] 20
--context_window [-w] 4 6
--active_site_offset_5 [-5] "-4"
--active_site_offset_3 [-5] "-4"
All IUPAC ambiguity codes are allowed and will be automically expanded, e.g. NGG -> AGG, TGG, CGG, GGG. Note that context_window[0] extends the sequence in the 5' direction, context_window[1] in the 3' direction. active_site_offsets are calculated relative to PAM-protospacer position, and should be passed in quotes if they are negative.
For Cas12A guide selection change generate_guides settings to
generate_guides \
-f <fasta> \
--pam TTTV --pam_5_prime -5 19 -3 23 -l 23 -w 7 2
Here the pam is 5-prime to the protospacer so --pam_5_prime flag is set and the length is increased 23. The window is resized for compatibility with DeepCpf1 and EnPAMGB scoring and final sequence should be 34 nts long.
For additional on-target scoring, including of Cas12A/Cpf1 guides, first install crisprScore (recommendation: install in a new conda environment). Once installed the crisprScore.R script can be used to score guides.
conda activate <crisprscore env>
./scripts/crisprscore.R
Usage: crisprscore.R <path_to_sgrna_bed_file> <method_number> <outputfile> [<additional settings> ... ]
Example: crisprscore.R tests/test_data/chr19_GRCm39_sgRNA.bed 2 Azimuth_scored_sgRNAs.bed
Methods:
1: RuleSet1 - SpCas9 (Length: 30)
2: Azimuth - SpCas9 (Length: 30)
3: DeepHF - SpCas9 (Length: 23)
4: Lindel - SpCas9 (Length: 65)
5: DeepCpf1 - AsCas12a (Length: 34)
6: EnPAMGB - enAsCas12a (Length: 34)
7: CRISPRscan - SpCas9 (Length: 35)
8: CasRx-RF - CasRx (Length: NA)
9: CRISPRai - SpCas9 (Length: 22)
10: CRISPRater - SpCas9 (Length: 20)
11: DeepSpCas9 - SpCas9 (Length: 30)
12: RuleSet3 - SpCas9 (Length: 30)
Additional optional argument for chunk size:
--chunk-size: Specify the size of chunks for processing the dataframe (optional)
Example with chunk size: crisprscore.R tests/test_data/chr19_GRCm39_sgRNA.bed 2 Azimuth_scored_sgRNAs.bed --chunk-size 10000
Additional settings for method 3 (DeepHF):
enzyme: Specify the enzyme (options: 'WT', 'ESP', 'HF')
promoter: Specify the promoter (options: 'U6', 'T7')
Example: crisprscore.R tests/test_data/chr19_GRCm39_sgRNA.bed 3 DeepHF_scored_sgRNAs.bed WT U6
Additional setting for method 5 (DeepCpf1):
--no-convertPAM: Specify whether non-canonical PAMs are converted to TTTC [default: TRUE]
Example: crisprscore.R tests/test_data/chr19_GRCm39_sgRNA.bed 5 DeepCpf1_scored_sgRNAs.bed --no-convertPAM
Additional setting for method 12 (RuleSet3):
--tracrRNA: Specify tracrRNA (options: 'Hsu2013', 'Chen2013')
Example: crisprscore.R tests/test_data/chr19_GRCm39_sgRNA.bed 12 rs3_scored_sgRNAs.bed Chen2013
The output of this script should be passed to score_guides in order to properly format for rank_guides. Additional score columns will be added unless the user specifies --skip_rs3 and/or --skip_gs2.
./scripts/crisprscore.R 34_sgrnas.bed 6 EnPAMGB_sgRNAs.bed
score_guides -b EnPAMGB_sgRNAs.bed --skip_rs3 --skip_gs2
Guidescan2 is not compatible with PAMs 5' to protospacers, for off-target scoring in these cases I suggest Flash Fry
Targeting noncoding elements can be guided by any NGS data that yields BED coordinate files. Additionally, a helper script bigwig_to_signalwindow.py can take a BED and BigWig signal file and return the window in each BED entry that has the highest mean signal.
preprocess_annotation
options:
-h, --help show this help message and exit
-g GTF, --gtf GTF GTF/GFF file to use for isoform filtering.
-t [TPM_FILES ...], --tpm_files [TPM_FILES ...]
A list of one or more isoform quantification files
produced by Salmon, Kallisto or FLAIR (FLAIR outputs
counts, not TPMs). The first column should contain
only the transcript_id and should exactly match the
transcript_ids in --gtf. All transcript_ids in each
TPM file must be common across all files and must be
found in the GTF file.
-f {salmon,kallisto,flair,mandalorian,infer}, --type {salmon,kallisto,flair,mandalorian,infer}
Specify TPM input type. 'infer' guesses the input type
based on the header line. [default: "infer"].
--mean MEAN For a given isoform, the mean tpm/count across samples
must be at least this to be considered, else discard
isoform. [default: 0.0]
--median MEDIAN For a given isoform, the median tpm/count across
samples must be at least this to be considered, else
discard isoform. [default: 0.0]
--min MIN For a given isoform, each sample must have at least
this tpm/count to be considered, else discard isoform.
[default: 0.0]
--max MAX For a given isoform, at least one sample must have at
least this tpm/count to be considered, else discard
isoform. [default: 0.0]
-n TOP_N, --top_n TOP_N
For a given gene, rank all isoforms by median_tpm,
keep the top_n ranked isoforms and discard the rest.
'-1' to keep all isoforms. [default: -1]
-c {median,mean,min,max}, --top_n_column {median,mean,min,max}
The metric by which to rank and filter top isoforms.
Used with '-n' to select expressed transcripts.
[default: median]
-m [{metagene,consensus,longest,shortest} ...], --model [{metagene,consensus,longest,shortest} ...]
Whether to output 'metagene', 'consensus', 'longest',
'shortest' model. 'longest' and 'shortest' select, for
a given gene, the transcript with the longest or
shortest CDS, for now noncoding genes are ignored.
Output is always after tpm filtering has been applied.
Multiple entries are allowed e.g. --model metagene
consensus longest [default: None]
-w TSS_WINDOW TSS_WINDOW, --tss_window TSS_WINDOW TSS_WINDOW
Pass two, space-separated, integers to specifiy the bp
window around the TSS as '<upstream>' '<downstream>'.
Strand-orientation is inferred, i.e. '<upstream>' will
be in the 5' direction of the TSS and <downstream> in
the 3' direction. e.g. --tss_window 250 150. [default:
None]
-e TES_WINDOW TES_WINDOW, --tes_window TES_WINDOW TES_WINDOW
Pass two, space-separated, integers to specifiy the bp
window around the transcription end site, TES, as
'<upstream>' '<downstream>'. Strand-orientation is
inferred, i.e. '<upstream>' will be in the 5'
direction of the TES and <downstream> in the 3'
direction. e.g. --tss_window 0 150. [default: None]
-x TX_TO_GENE, --tx_to_gene TX_TO_GENE
A TSV with transcript IDs in the first column and Gene
IDs in the second. The transcript IDs must match the
first column entries of the --quant_files. If this is
not provided it will be deduced from the GTF/GFF3 and
saved as
'./annotations/intermediateFiles/tx2gene.tsv'.
index_genome
options:
-h, --help show this help message and exit
-f FASTA, --fasta FASTA
FASTA file to use as a reference for index creation.
-o OUTPUT_PREFIX, --output_prefix OUTPUT_PREFIX
Prefix for output files. [default='gscan_index']
--locations_to_keep [LOCATIONS_TO_KEEP ...]
List of BED/GTF files with coordinates to use for
index creation. These locations will be used for off-
target scoring. If multiple files are passed,
coordinates will be merged with a union operation.
Leave empty to use entire fasta.
--feature FEATURE For any GTF/GFF in '--locations_to_keep', only this
feature will be used for determining appropriate
sgRNA. The feature should match an entry in the third
column of the GTF/GFF. [default: 'transcript']
-w CONTEXT_WINDOW CONTEXT_WINDOW, --context_window CONTEXT_WINDOW CONTEXT_WINDOW
Pass two, space-separated, integers to specifiy the
nucleotide window around the --locations_to_keep
'<upstream>' '<downstream>'. This can be used to
expand the window around the final intervals e.g. '-w
1000 1500' expands chr1 2000 3500 -> chr1 1000 5000
Good for CRISPRi/a [default: 20 20]
generate_guides
options:
-h, --help show this help message and exit
-f FASTA, --fasta FASTA
FASTA file to use as a reference for sgRNA generation.
-p PAM, --pam PAM Protospacer adjacent motif to match. All IUPAC
ambiguity codes are accepted as well as standard ATCG.
[default: NGG]
-l SGRNA_LENGTH, --sgRNA_length SGRNA_LENGTH
Length of sgRNA to generate. [default: 20]
-w CONTEXT_WINDOW CONTEXT_WINDOW, --context_window CONTEXT_WINDOW CONTEXT_WINDOW
Pass two, space-separated, integers to specifiy the
nucleotide window around the sgRNA as '<upstream>'
'<downstream>'. This can be used for downstream
scoring, For Ruleset3 use -w 4 6 to obtain an
appropriate score context. [default: 4 6]
-5 ACTIVE_SITE_OFFSET_5, --active_site_offset_5 ACTIVE_SITE_OFFSET_5
Where cut occurs relative to PAM 5' end. To avoid
error, use '=' sign when passing a negative number,
e.g. --active_site_offset_5=-1 [default: -4]
-3 ACTIVE_SITE_OFFSET_3, --active_site_offset_3 ACTIVE_SITE_OFFSET_3
Where cut occurs relative to PAM 5' end. [default: -2]
To avoid error, use '=' sign when passing a negative
number, e.g. --active_site_offset_3=-3 [default: -4]
-k [LOCATIONS_TO_KEEP ...], --locations_to_keep [LOCATIONS_TO_KEEP ...]
List of BED/GTF files with coordinates in which the
sgRNA desired. If the sgRNA cutsite does not intersect
coordinates in these files they are discarded. Leave
blank to keep all sgRNA. e.g. atac_peak.bed genes.gtf
--feature FEATURE For any GTF/GFF in '--locations_to_keep', only this
feature will be used for determining appropriate
sgRNA. The feature should match an entry in the third
column of the GTF/GFF. [default: 'exon']
--join_operation {merge,intersect}
How to treat '--locations_to_keep' if multiple files
are passed. Either 'merge' or 'intersect' can be used
and work as described in Bedtools. If 'merge', sgRNA
will be kept if its cutsite intersects an entry in ANY
of the files, if 'intersect' the cutsite must
intersect an entry in EACH file. [default:
'intersect']
--locations_to_discard [LOCATIONS_TO_DISCARD ...]
List of BED/GTF files with coordinates where sgRNA
should not target. If the sgRNA cutsite intersects
coordinates in these files the sgRNA is discarded.
Leave blank to keep all sgRNA. e.g. TSS.bed
coding_genes.gtf
--prefix PREFIX Prefix to use for sgRNA identifiers. [default: None]
--gc_range GC_RANGE GC_RANGE
Pass two, space-separated, integers to specifiy the
percentile range of GC content e.g. '--gc_range 25
75'. [default: 0 100]
--discard_poly_T Whether to discard polyT (>TTT) sgRNA. Recommend True
for PolIII promoters [default: False]
--discard_poly_G Whether to discard polyT (>GGGG) sgRNA. [default:
False]
--restriction_patterns [RESTRICTION_PATTERNS ...]
Reject sgRNA with these restriction patterns. Also
checks 5'flank+sgRNA+3'flank, and reverse complement,
if provided. For multiple values, separate by space.
e.g. GCGGCCGC TCTAGA CACCTGC
--flank_5 FLANK_5 include the 5' context of the lentivirus vector. Used
in conjunction with --restriction_patterns to remove
incompatible sgRNA
--flank_3 FLANK_3 include the 3' context of the lentivirus vector. Used
in conjunction with --restriction_patterns to remove
incompatible sgRNA
--min_chr_length MIN_CHR_LENGTH
Minimum chromosome length to consider for sgRNA
generation. [default: 20]
--pam_5_prime If the PAM is positioned 5' to the protospacer set
this flag, e.g. for Cas12a sgRNAs [default: False]
--coords_as_active_site
Whether to output bed coordinates at the active site
rather than the coordinates of the entire protospacer.
For purposes of keeping or discarding sgRNAs, overlap
with the active site coordinates will be used
regardless [default: True]
-o OUTPUT_PREFIX, --output_prefix OUTPUT_PREFIX
Prefix for output files.
-t THREADS, --threads THREADS
Number of threads. [default: 4]
score_guides
options:
-h, --help show this help message and exit
-b SGRNA_BED, --sgrna_bed SGRNA_BED
sgrnas.bed ouput of GenerateGuides.
-i [GUIDESCAN2_INDICES ...], --guidescan2_indices [GUIDESCAN2_INDICES ...]
One or more, space-separate Guidescan2 indices. A
specificity score will be calculated against each
index separately.
--tracr {Hsu2013,Chen2013,both}
TracrRNA version for cleavage scoring. Either
'Hsu2013' or 'Chen2013' or 'both', see
https://github.com/gpp-rnd/rs3 for details.
-t THREADS, --threads THREADS
Number of threads [default: 8]
--threshold THRESHOLD
Threshold for Guidescan2 off-target hits. If off-
targets are found this distance away the sgRNA will be
discarded, i.e. set to 2 to discard any guides with a
0, 1 or 2 mismatches from another PAM adjacent
sequence. --threshold=-1 to retain all guides
[default: 2]
--mismatches MISMATCHES
Number of mismatches for Guidescan2 off-target scoring
[default: 3]
--rna_bulges RNA_BULGES
RNA bulges for Guidescan2 off-target scoring [default:
0]
--dna_bulges DNA_BULGES
DNA bulges for Guidescan2 off-target scoring [default:
0]
--mode {succinct,complete}
Whether Guidescan2 temporary output should be succinct
or complete mode [default: 0]
--alt_pams [ALT_PAMS ...]
One or more, space-separate alternative pams for off-
target consideration. e.g. NAG
-d, --drop_duplicates
Drop exact duplicate sgRNAs before scoring to save
time. Set flag to retain duplicates. [default: True]
--skip_rs3 Set flag to skip RS3 scoring [default: False]
--skip_gs2 Set flag to skip Guidescan2 scoring [default: False]
--min_rs3 MIN_RS3 Minimum cleavage RS3 score. RS3 cleavage scores are
formatted as z-scores, so this is interpreted as a
standard deviation cutoff. Functionality also
available in rank_guides.py. Applying at this stage
can increase speed by filtering before off-target
scoring. [default: None]
--chunk_size CHUNK_SIZE
Number of sgRNAs to hold in memory for cleavage
scoring and off-target filtering. Reduce if memory
constrained. Increasing may improve runtime [default:
100000]
-o OUTPUT_PREFIX, --output_prefix OUTPUT_PREFIX
Prefix to use for files.
-k, --keep_tmp Set flag to keep temporary Guidescan2 output [default:
False]
rank_guides
options:
-h, --help show this help message and exit
-k SCORED_GUIDES, --scored_guides SCORED_GUIDES
<score_guides_output>.tsv output from score_guides.
-t TARGETS, --targets TARGETS
BED/GTF/GFF used to select final guides per target.
For GTF/GFF, set --target_mode to either 'gene' or
'transcript'. For BED, targets are each entry. Use '--
number_of_targets' to set the number of guides chosen
for each target.
--target_mode {gene,tx}
If a GTF/GFF is used to select targets, sgRNAs can be
grouped at either the 'tx' or 'gene' level e.g. '--
target_mode gene -n 10' chooses 10 guides per gene, '
--target_mode tx -n 10' chooses 10 per transcript
[default: gene].
-f FEATURE, --feature FEATURE
If GTF/GFF passed, use this feature for processing
e.g. 'exon', 'CDS', '5UTR', etc. The feature appears
in the third column of the GTF/GFF [default: CDS].
-p PERCENTILE_RANGE PERCENTILE_RANGE, --percentile_range PERCENTILE_RANGE PERCENTILE_RANGE
Allowable range of guide for each transcript and
feature set, e.g. '-p 60 80 -f exon' returns sgRNAs in
the 60th to 80th percentile of exons for a given
transcript. Default setting returns guides anywhere in
the CDS for each transcript [default: 0 100]
-n NUMBER_OF_GUIDES, --number_of_guides NUMBER_OF_GUIDES
Number of guides returned per target.'-1' to keep all
guides [default: -1]
--min_spacing MIN_SPACING
The minimum nucleotide space between guides for a
given target. e.g. --min_spacing 10, requires guides
10 nts appart. 0 to allow overlapping guides.[default:
0]
--output_all Set flag to save sgRNA-target TSVs at each stage of
filtering rather than just the end.[default: False]
--plot_histogram Set flag to plot a histogram of the distribution of
sgRNAs per target after each filtering step. Sets '--
output_all' to True.[default: False]
-c [FILTERING_COLUMNS ...], --filtering_columns [FILTERING_COLUMNS ...]
One or more space-separated column names used for
filtering. Uses raw values. e.g. '-c rs3_z_score
specificity_Hg38_index'.
-m [MINIMUM_VALUES ...], --minimum_values [MINIMUM_VALUES ...]
A space-separated list of minimum values for each
column in passed by --ranking_columns. e.g. '-c
rs3_z_score specificity_Hg38_index -m "-1" 0.2'
Default is no minimum [default: None]
-r [RANKING_COLUMNS ...], --ranking_columns [RANKING_COLUMNS ...]
One or more space-separated column names used for
guide ranking. e.g. '-r rs3_score_Hsu2013
rs3_score_Chen2013'.
-w [COLUMN_WEIGHTS ...], --column_weights [COLUMN_WEIGHTS ...]
A space-separated list of weight values for each
column in passed by --ranking_columns. e.g. '-c
rs3_score specificity_Hg38_index -w 1 0' Default is
equal weighting for all ranking columns.
--normalize_columns Scale ranking column values to 0 to 1 [default: True]
-o OUTPUT_PREFIX, --output_prefix OUTPUT_PREFIX
Prefix for output file