Assessing the benefit of pre-training a thought classification model using neural prediction with an LSTM. Read the paper.
I perform self-supervised training (LeCun & Misra, 2021) to pre-train a machine learning model using the LSTM architecture (Hochreiter & Schmidhuber, 1997) on functional near-infrared spectroscopic (fNIRS) neuroimaging data (Naseer & Hong, 2015) from the NIRx NIRSport2 system (NIRx, 2021) and transfer and fine-tune it for a BCI thought classification task (Yoo et al., 2018) as is done with language models (C. Sun et al., 2019). As far as I am aware, this is the first example of such work.
Accompanies a YouTube series.
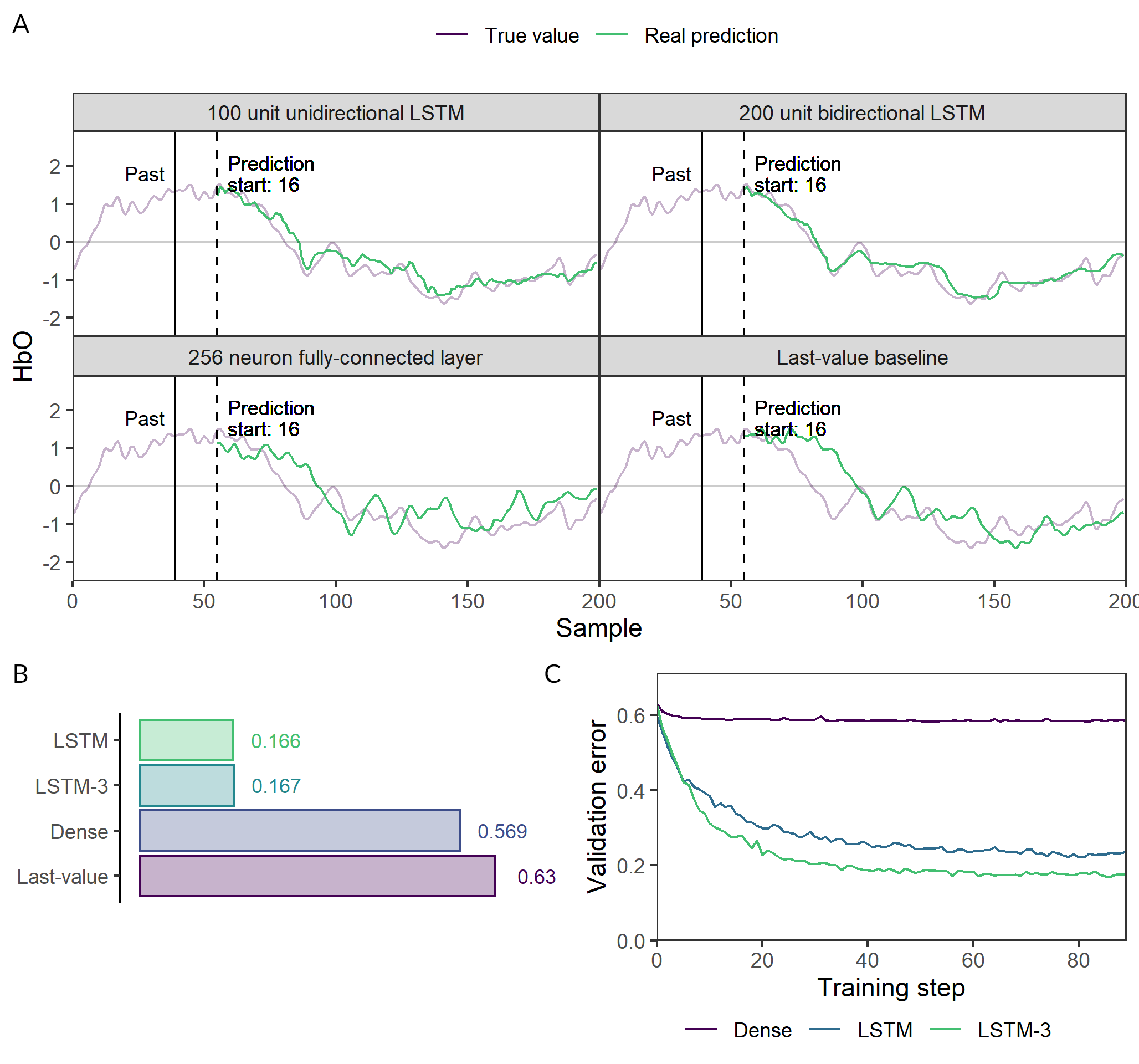
The 1-layer LSTM, 3-layer LSTM and dense pre-trained models were trained to predict the brain activity in channel 1 4.2 seconds in the future given 10 seconds of data. These were highly succesful and LSTMs performed much better than the fully-connected and the designed baseline (fig. 3).
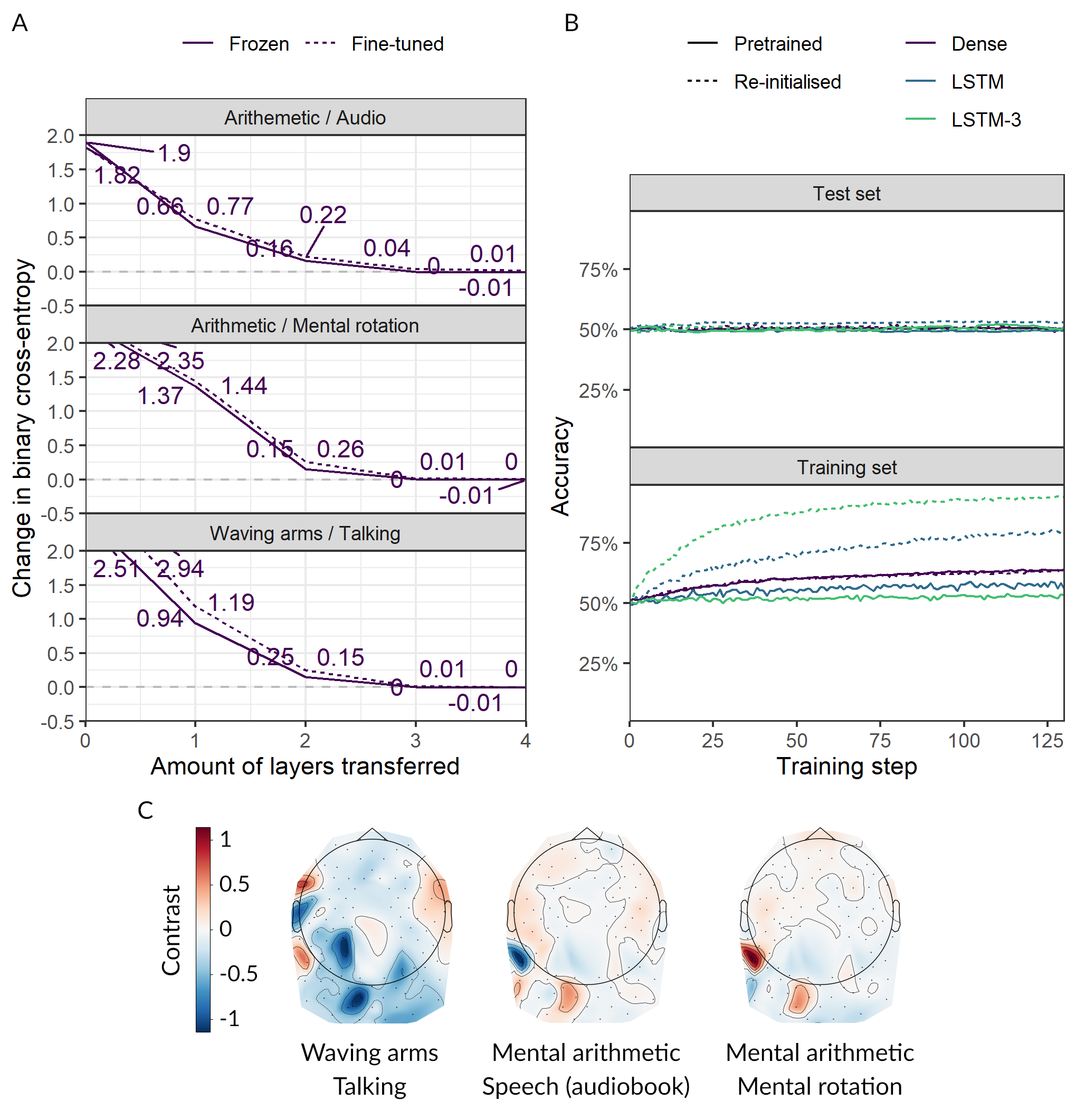
The model weights were transferred and the last layer was replaced with a 256 dense layer and a sigmoid binary classifier. These underfit horribly but the pre-training avoided extreme overfitting (fig. 4).
| File name | Description |
|---|---|
exp_train_st_all.py |
👩🔬 Trains the neural prediction models with basis in a configuration dictionary in the script. To run this, you need to connect W&B. |
exp_bci_task.py |
👩🔬 Trains the classification models. Also has configurations and need a login to W&B. **Run generate_augmented_datasets.py](code/generate_augmented_datasets.py)** to generate augmented datasets in [data/datasets` before running this. |
experiment_bci.py |
👩🔬 Code for running the terminal experimental paradigm. Starts an LSL stream that logs triggers in the .snirf fNIRS output files. |
helper_functions.py |
👩💻 An extensive selection of helper functions generally referred to by .py code in this directory. |
generate_augmented_datasets.py |
👩💻 Generates .npy train/test datasets with/without augmentation to use for exp_bci_task.py. |
3_data_figure.py |
📊 Generates prediction data for figure 3A. |
4_brain_plot.py |
📊 Generates contrast brain plot in figure 4C. |
data_wandb.py |
📊 Collects data from W&B using their api. Also requires you to login. |
figures.Rmd |
📊 Generates figures from the data collected from the above scripts. Each figure can be run isolated in their own code chunk and outputs to media/figures. |
analysis.Rmd |
✍ Simple analyses and unstructured code. |
pipeline_math.Rmd |
✍ Goes through an unstructured explanation of the math implemented in R. |
- Data
- Analysis: Datasets used in
analysis.rmd,figures.rmdandpipeline_math.rmd - Datasets: Generated train/test datasets as
.npyfromgenerate_augmented_datasets.py(excluded by.gitignore) - Snirf: The raw fNIRS data files (
.snirf) - Visualization: Two datasets exclusively used for visualization in
figures.rmd - Weights: Randomly initialized model weights to replace layer weights when loading pre-trained models
- Analysis: Datasets used in
- Media: Misc. image ouputs and showcases
- Figures: All figures and subfigures used in the paper, editable with
figures.xd
- Figures: All figures and subfigures used in the paper, editable with
- Models: Contains the pre-trained models used in the paper's transfer learning part