This repository holds materials that were used at the Biosignals 2023 tutorial as well as general learning material.
Recommended Integrated Development Environment (IDE): VSCode
Alternative IDEs: Pycharm, IDLE etc.
Recommended Package Manager: Conda/Anaconda
Alternative Package Managers: Pip(not recommended), Mamba (will take effort and lots of knowledge)
Version control (VC): Git
Alternative VC: existing VC was done in Git so the only alternative at this point we can imagine is Perforce (not recommended because nobody in the core teams will know how to help you, good thing you are a genius)
Binders and other resources for learning the basic computational skills needed to understand our work.
For a practical beginner introduction to Python, we reccomend the software carpentry on Python. You should also pick up version control as early as possible, we reccomend this carpentry tutorial. For some tips on coding on our project, see our best_practices.md file .To learn notebook navigation, as well as more specific tips about ReSurfEMG we reccomend using our Jupyter binders (just click on the badge links like this one: ). Did you start tinkering somewhere and make a mistake? See our fixing_mistakes.md file.
We presented a tutorial at Biosignals 2023 in February 2023, the materials are here.
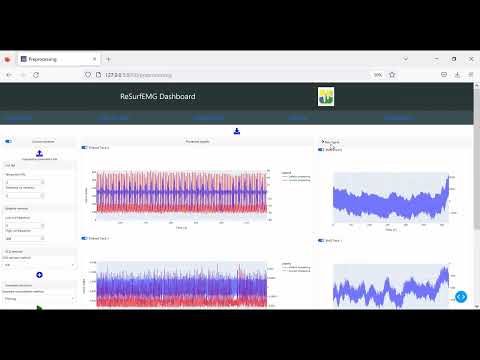
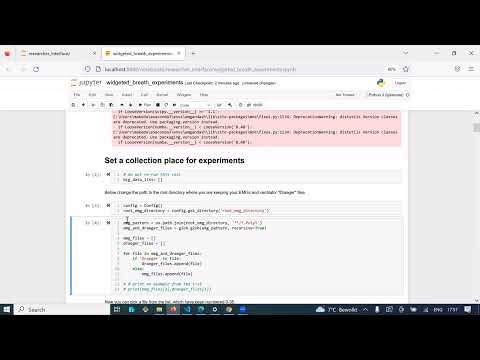
Video demonstrations: to see videos click the links below (more on the way)
About the researcher interface:
✨Copyright 2022 Netherlands eScience Center and U. Twente Licensed under the Apache License, version 2.0. See LICENSE for details.✨