NeSVoR is a package for GPU-accelerated slice-to-volume reconstruction.
This package is the accumulation of the following works:
[1] SVoRT: Iterative Transformer for Slice-to-Volume Registration in Fetal Brain MRI (Springer | Arxiv)
[2] NeSVoR: Implicit Neural Representation for Slice-to-Volume Reconstruction in MRI (IEEE|TechRxiv)
NeSVoR is a deep learning package for solving slice-to-volume reconstruction problems (i.e., reconstructing a 3D isotropic high-resolution volume from a set of motion-corrupted low-resolution slices) with application to fetal/neonatal brain MRI, which provides
- Motion correction by mapping 2D slices to a 3D canonical space using Slice-to-Volume Registration Transformers (SVoRT).
- Volumetric reconstruction of multiple 2D slices using implicit neural representation (NeSVoR).
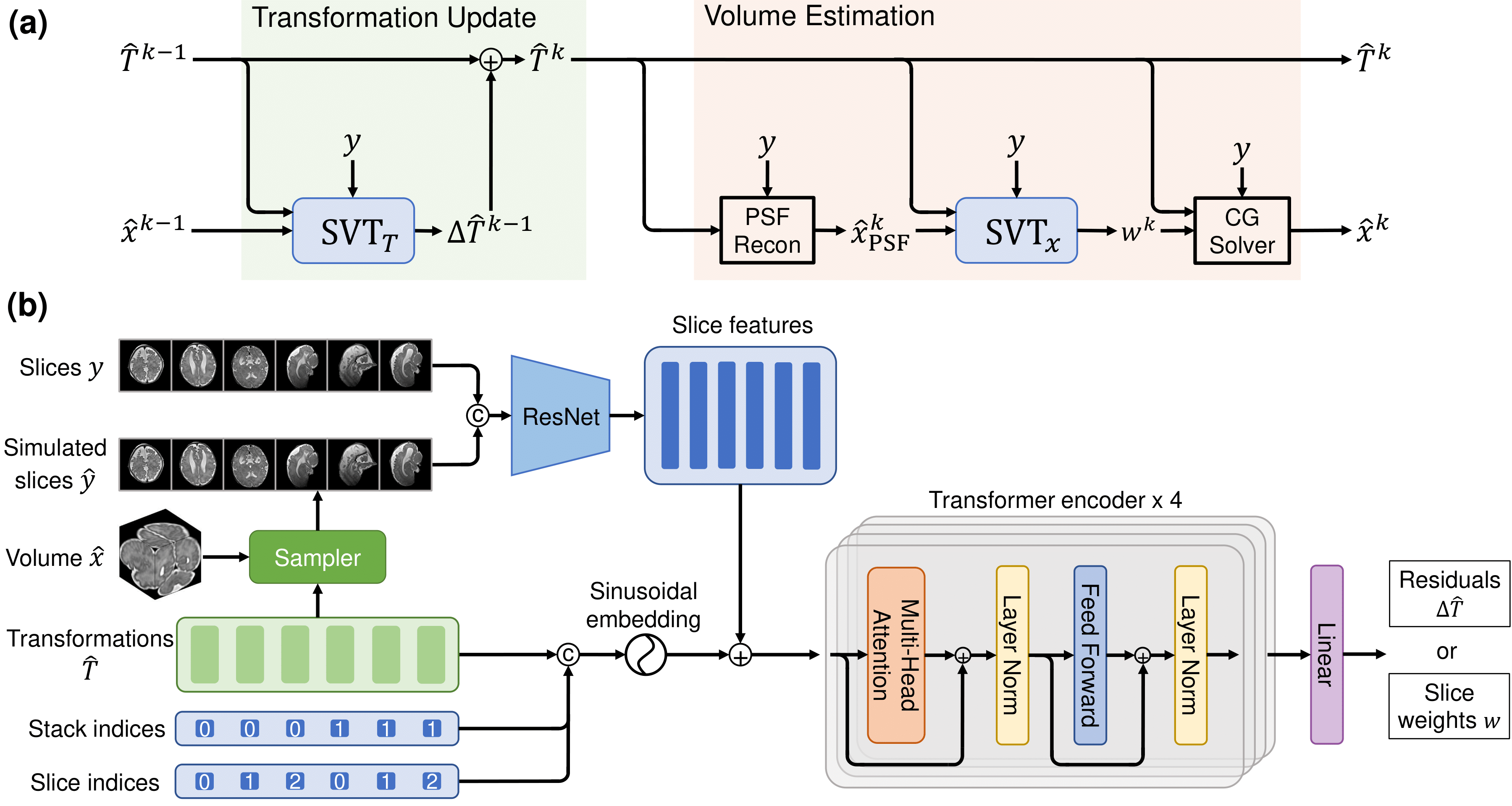
Figure 1. SVoRT: an iterative Transformer for slice-to-volume registration. (a) The k-th iteration of SVoRT. (b) The detailed network architecture of the SVT module.
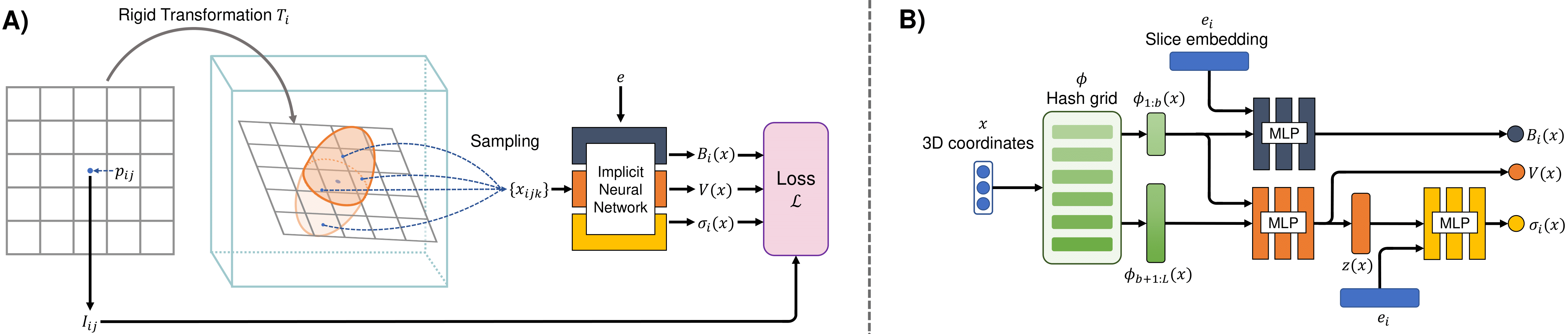
Figure 2. NeSVoR: A) The forward imaging model in NeSVoR. B) The architecture of the implicit neural network in NeSVoR.
If you are installing from source, you will need:
- An NVIDIA GPU
- Python 3.8 or later;
- GCC/G++ 7.5 or higher;
- CUDA v10.2 or higher;
- CMake v3.21 or higher.
see tiny-cuda-nn for more on prerequisites
git clone https://github.com/daviddmc/NeSVoR
cd NeSVoR
pip install -r requirements.txt
Install PyTorch extension of tiny-cuda-nn. Make sure the installed CUDA version mismatches the version that was used to compile PyTorch. Then, run the following command (see this for more details)
pip install git+https://github.com/NVlabs/tiny-cuda-nn/#subdirectory=bindings/torch
pip install .
You may follow this guide to install Docker and NVIDIA Container Toolkit
docker pull junshenxu/nesvor:v0.1.0
docker run --gpus '"device=0"' -it junshenxu/nesvor:v0.1.0
Note: our image was built with CUDA 11.6.
NeSVoR currently supports the following commands.
nesvor reconstruct: reconstruct a 3D volume (i.e., train a NeSVoR model) from either multiple stacks of slices (NIfTI) or a set of motion-corrected slices (the output of register).
nesvor register: register stacks of slices using a pretrained SVoRT model.
nesvor sample-volume: sample a volume from a trained NeSVoR model.
nesvor sample-slices: simulate slices from a trained NeSVoR model.
run nesvor <command> -h for a full list of parameters of a command.
The reconstruct command can be used to reconstruct a 3D volume from N stacks of 2D slices (in NIfTI format, i.e. .nii or .nii.gz).
A basic usage of reconstruct is as follows, where mask-i.nii.gz is the ROI mask of the i-th input stack.
nesvor reconstruct \
--input-stacks stack-1.nii.gz ... stack-N.nii.gz \
--stack-masks mask-1.nii.gz ... mask-N.nii.gz \
--output-volume volume.nii.gz
A more elaborate example could be
nesvor reconstruct \
--input-stacks stack-1.nii.gz ... stack-N.nii.gz \
--stack-masks mask-1.nii.gz ... mask-N.nii.gz \
--thicknesses <thick-1> ... <thick-N> \
--output-volume volume.nii.gz \
--output-resolution 0.8 \
--output-model model.pt \
--weight-image 1.0 \
--image-regularization edge \
--delta 0.2 \
--n-iter 5000 \
--level-scale 1.38 \
--coarsest-resolution 16.0 \
--finest-resolution 0.5 \
--n-levels-bias 0 \
--n-samples 128
Run nesvor reconstruct --h to see the meaning of each parameter.
Given multiple stacks at inputs, reconstruct first corrects the motion in the input stacks using SVoRT (the same as what register did), and then trains a NeSVoR model that implicitly represent the underlying 3D volume, from which a discretized volume (i.e., a 3D tensor) can be sampled.
reconstruct can also take a folder of motion-corrected slices as inputs.
nesvor reconstruct \
--input-slices <path-to-slices-folder> \
--output-volume volume.nii.gz
This enables the separation of registration and reconstruction. That is, you may first run register to perform motion correction, and then use reconstruct to reconstruct a volume from a set of motion-corrected slices.
register takes mutiple stacks of slices as inputs, performs motion correction, and then saves the motion-corrected slices to a folder.
nesvor register \
--input-stacks stack-1.nii.gz ... stack-N.nii.gz \
--stack-masks mask-1.nii.gz ... mask-N.nii.gz \
--registration <method> \
--output-slices <path-to-save-output-slices>
register currently supports the following methods:
svort: the full SVoRT model;svort-stack: only apply stack transformations of SVoRT;stack: stack-to-stack rigid registration;none: no registration.
Upon training a NeSVoR model with the reconstruct command, you can sample a volume at arbitrary resolutions with the sample-volume command.
nesvor sample-volume \
--output-volume volume.nii.gz \
--input-model model.pt \
--output-resolution 0.5
You may sample slices from the model using the sample-slices command. For each slice in <path-to-slices-folder>, the command simulates a slice from the NeSVoR model at the corresponding slice location.
nesvor sample-slices \
--input-slices <path-to-slices-folder> \
--input-model model.pt \
--simulated-slices <path-to-save-simulated-slices>
For example, after running reconstruct, you can use sample-slices to simulate slices at the motion-corrected locations and evaluate the reconstruction results by comparing the input slices and the simulated slices.
SVoRT
@inproceedings{xu2022svort,
title={SVoRT: Iterative Transformer for Slice-to-Volume Registration in Fetal Brain MRI},
author={Xu, Junshen and Moyer, Daniel and Grant, P Ellen and Golland, Polina and Iglesias, Juan Eugenio and Adalsteinsson, Elfar},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={3--13},
year={2022},
organization={Springer}
}
NeSVoR
@article{10015091,
author={Xu, Junshen and Moyer, Daniel and Gagoski, Borjan and Iglesias, Juan Eugenio and Ellen Grant, P. and Golland, Polina and Adalsteinsson, Elfar},
journal={IEEE Transactions on Medical Imaging},
title={NeSVoR: Implicit Neural Representation for Slice-to-Volume Reconstruction in MRI},
year={2023},
volume={},
number={},
pages={1-1},
doi={10.1109/TMI.2023.3236216}
}