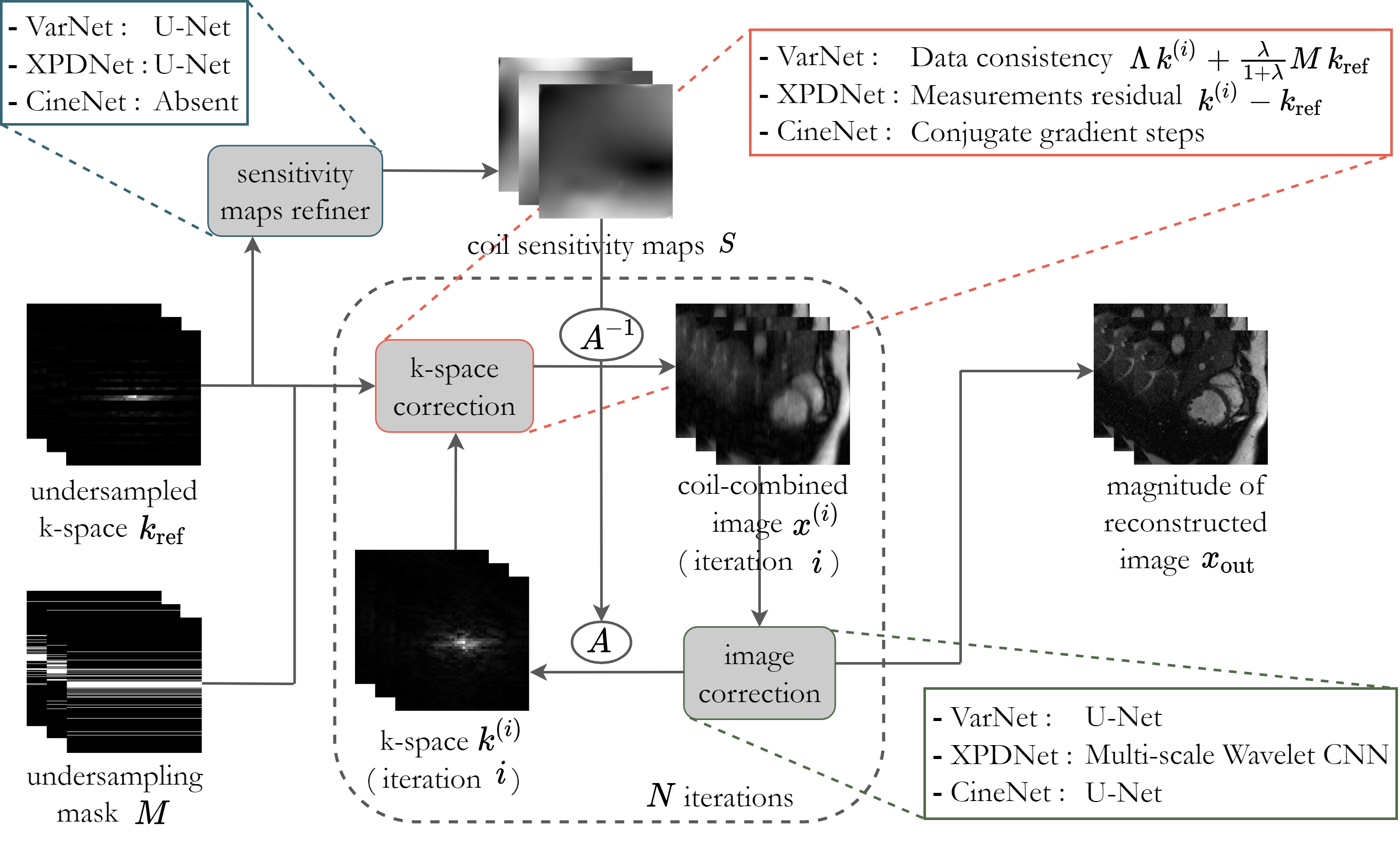
This project presents novel methods to tackle the problem of dynamic MRI reconstruction (2D space + 1D time) of accelerated multi-coil cardiac data.
The proposed methods are inspired by the recent deep-learning models
that achieved state-of-the-art results in the 2020 FastMRI challenge for static MRI reconstruction of the brain and the knee, and by
- CineNet [3]
for dynamic reconstruction of 2+1D cardiac data.
The underlying architecture for all proposed models is based on the cross-domain network with unrolled optimisation. A high-level diagram of the architecture is shown below:
The dynamic aspect of the MRI data is taken into account by adjusting the models to process 2+1D data volumes and exploit the inherent temporal redundancies of cine cardiac data. To this aim, each model was structured including five dynamic variants:
- 2D - static scenario, each temporal slice is processed independently
- 3D - each 2+1D volume is processed as a whole; model is isotropic towards each dimension
- XT - two copies of each volume are rotated in the two 2D spatio-temporal domains (x-t plane and y-t plane), respectively, then processed by separate image correction networks and finally recombined
- XF - same as for 'XT', but volume firstly undergoes an FFT along the temporal dimension to sparsify the domain
- CRNN - leverage the time-dependency directly by implementing a recurrence along the temporal direction and also across the unrolled iterations. This variant is based on the Convolutional Recurrent Neural Network introduced by [4]
- Clone the repository:
git clone https://github.com/f78bono/deep-cine-cardiac-mri.git
- Create a new virtual environment and install the requirements in
./requirements.txt - Set up a BART environment following the instruction in this notebook
- Change the paths in the following files according to your root directory:
# traintest_scripts/dirs_path.yaml
data_path: /path/to/data
log_path: /root/traintest_scripts
save_path: /root/results
# reconstruction/pl_modules/mri_module
path_config = pathlib.Path("/root/traintest_scripts/dirs_path.yaml")
# traintest_scripts/model/train_test_model.py
path_config = pathlib.Path("/root/traintest_scripts/dirs_path.yaml")
The data set used in this project consists of breath-hold, retrospectively cardiac-gated, Cartesian, bSSFP multi-coil cine cardiac raw data. Each MRI volume is stored in a HDF5 file that contains the raw k-space data, with dimensions (number of slices, height, width, number of coils).
The path to the data directory expects subdirectories train/valid/test. If a
subdirectory inference is also included, the program can be given the option
to store the model outputs for visualisation purposes. The code for dealing with
the raw HDF5 files can be found in ./reconstruction/data/mri_data.py. Adjust the
following settings according to the specifics of the data set used:
scaling = 1e6
crop_shape = (200, 200)
crop_target = (180, 180)
n_slices = 15
filter_size = [0.7, 0., 0.3, 0.3]
The scripts for training and testing each model can be found in the traintest_scripts
subfolders. Running a script with no arguments, for example
python3 train_test_varnet.py
will start the training of the model (in this case XF-VarNet) with default parameters. Pass optional arguments to overwrite the default parameters, for example
python3 train_test_varnet.py \
--epochs 50 \
--save_checkpoint 1 \
--num_cascades 6 \
--dynamic_type CRNN
For testing use the command
python3 train_test_varnet.py --mode test --load_model 1
plus the arguments needed for the model under consideration.
A detailed description of each argument can be found in the source code.
A qualitative assessment of the models was perfomed by direct observation of the reconstructed output along with the absolute error between normalised target and output. Check this notebook for a visual comparison of the different model outputs.
The quantitative metrics used for evaluation are:
- Structural similarity index measure (SSIM)
- Normalised mean square error (NMSE)
- Peak signal-to-noise ratio (PSNR)
These statistics are compiled automatically during testing and stored in the results
folder.
[1] Sriram, Anuroop et al. (2020). "End-to-End Variational Networks for Accelerated MRI Reconstruction". In: Medical Image Computing and Computer Assisted Intervention - MICCAI 2020. Cham: Springer International Publishing, pp. 64 - 73.
[2] Ramzi, Zaccharie et al. (2021). "XPDNet for MRI Reconstruction: an application to the 2020 fastMRI challenge". In: arXiv: 2010.07290.
[3] Kofler, Andreas et al. (2021). "An end-to-end-trainable iterative network architecture for accelerated radial multi-coil 2D cine MR image reconstruction". In: Medical Physics 48.5, pp. 2412 - 2425.
[4] Qin, Chen et al. (2019). "Convolutional Recurrent Neural Networks for Dynamic MR Image Reconstruction". In: IEEE Transactions on Medical Imaging 38.1, pp. 280 - 290.