A python code for matching points on the sphere using healpix.
This code is about 3.5 times faster than HTM in the esutil library.
# For quick matches, use the match() function
import smatch
nside=4096 # healpix nside
maxmatch=1 # return closest match
# ra,dec,radius in degrees
matches = smatch.match(ra1, dec2, radius, ra2, dec2,
nside=nside, maxmatch=maxmatch)
# in the above call, radius can be a scalar or the
# same size as ra1,dec1
# the output matches structure holds the indices of the matches from each data
# set, and the cosine of the distance between them
print(m.dtype.descr)
[('i1', '<i8'), ('i2', '<i8'), ('cosdist', '<f8')]
# access via the indices. These should match up one to one
ra1matched = ra1[ matches['i1'] ]
dec1matched = dec1[ matches['i1'] ]
ra2matched = ra2[ matches['i2'] ]
dec2matched = dec2[ matches['i2'] ]
# set maxmatch=0 to get all matches, set to a positive
# integer to return that many closest matches
all_matches = smatch.match(ra1, dec2, radius, ra2, dec2,
nside=nside, maxmatch=0)
some_matches = smatch.match(ra1, dec2, radius, ra2, dec2,
nside=nside, maxmatch=3)
# you can also match a catalog to itself, ignoring exact
# matches
some_matches = smatch.match_self(ra, dec, radius)
# A more flexibile interface is a Catalog. For example it can
# be used to match the same data set to multiple other data sets
cat=smatch.Catalog(ra1, dec1, radius, nside=nside)
print(cat)
smatch catalog
nside: 512
pixel area (sq deg): 0.013114
npoints: 100
cat.match(ra2, dec2, maxmatch=maxmatch)
cat.match(ra3, dec3, maxmatch=maxmatch)
print("found:",cat.nmatches,"matches")
matches = cat.matches
# matching the catalog to itself, ignoring exact matches
cat.match_self(maxmatch=maxmatch)
#
# Writing matches to file
#
# This useful if the number of matches is large, and cannot be
# held in memory
# using the convenience function
fname="matches.dat"
smatch.match(ra1, dec2, radius, ra2, dec2, file=fname)
smatch.match_self(ra, dec, radius, file=fname)
# using a catalog
cat.match(fname, ra3, dec3, file=fname)
cat.match_self(file=fname)
# you can read them later
matches=smatch.read_matches(fname)
# if the matches are too large to read, you can use packages
# such as these to read subsets
# recfile: https://github.com/esheldon/recfile
# esutil.recfile: https://github.com/esheldon/esutil
# (the esutil version is a copy of recfile)
from esutil.recfile import Recfile
dtype=smatch.smatch_dtype
# read 1000 elements starting at 10000
start=10000
end=11000
with Recfile(filename, "r", dtype=dtype, delim=' ') as robj:
data=robj[start:end]All unit tests should pass
import smatch
smatch.test()
testCreate (smatch.test.TestSMatch) ... ok
testMatch (smatch.test.TestSMatch) ... ok
testMatch2File (smatch.test.TestSMatch) ... ok
----------------------------------------------------------------------
Ran 3 tests in 0.003s
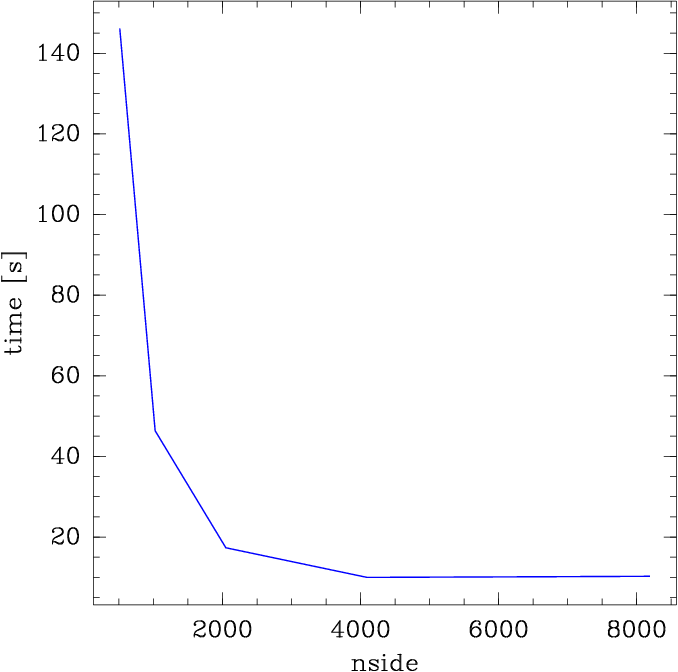
OKFor catalogs with object density of the Dark Energy Survey, searching within 10
arcseconds, higher nside results in faster search times, with some tradeoff
in memory usage. 4096 or 2048 are probably sufficient for this use case.
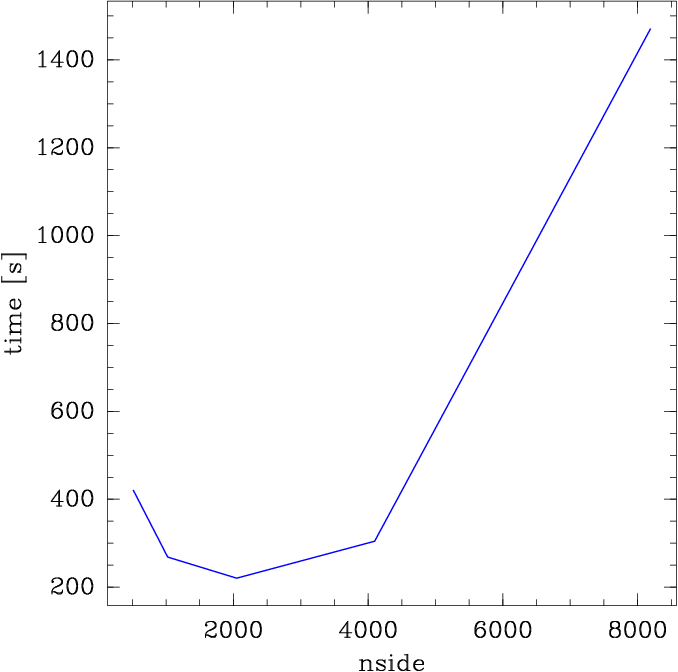
For larger search radii, smaller nside, and thus larger area pixels, works better; for example with the same catalog and a 200 arcsec search radius, nside=2048 is faster.