python学习在线资源:Cookbook for python
Basic-Machine-Learning-Algorithms
Memorial Sloan Kettering Cancer Center (MSK)OncoKB: A Precision Oncology Knowledge Base
命名标准化HGNC:https://www.genenames.org
GeneCards:https://www.genecards.org
文档转化Pandoc a universal document converter
表格转换工具:markdown-json-reStructuredText
- pandoc-markdown2pdf文档转化
- UCSC
- GATK
- Linux
- python3
- R语言
- Metagenomic宏基因组
- micro微生物基因组
- Benchmark_SNV_CNV_SV
- sample_qc
- SAM_VCF
- somatic
- germline
- ACMG_AMP_SCCO_CAP
- silico数据模拟
- Genome in a Bottle Consortium Genomes
- Mac电脑
- low pass WGS
- single cell单细胞
- small RNA
- methylation
- TMB_MSI
- HRD
- T2T-CHM13_GRCh38
GA4GH/GIAB stratifications: https://github.com/genome-in-a-bottle/genome-stratifications
Genome in a Bottle Consortium: http://www.genomeinabottle.org/
gnomAD: https://gnomad.broadinstitute.org/
Human Pangenome Reference Consortium: https://humanpangenome.org/
T2T-CHM13: https://github.com/marbl/CHM13
My Cancer Genome:www.mycancergenome.org
JAX Clinical Knowledgebase:https://ckb.jax.org
Clinical Interpretation of Variants in Cancer:https://civic.genome.wustl.edu
Oncology Knowledge Base:https://oncokb.org
Clinical Genome:https://clinicalgenome.org
COSMIC: https://cancer.sanger.ac.uk/cosmic
HGMD: http://www.hgmd.cf.ac.uk/ac/index.php
ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/
1000 Genomes: https://www.internationalgenome.org/
GWAS Catalog: https://www.ebi.ac.uk/gwas/
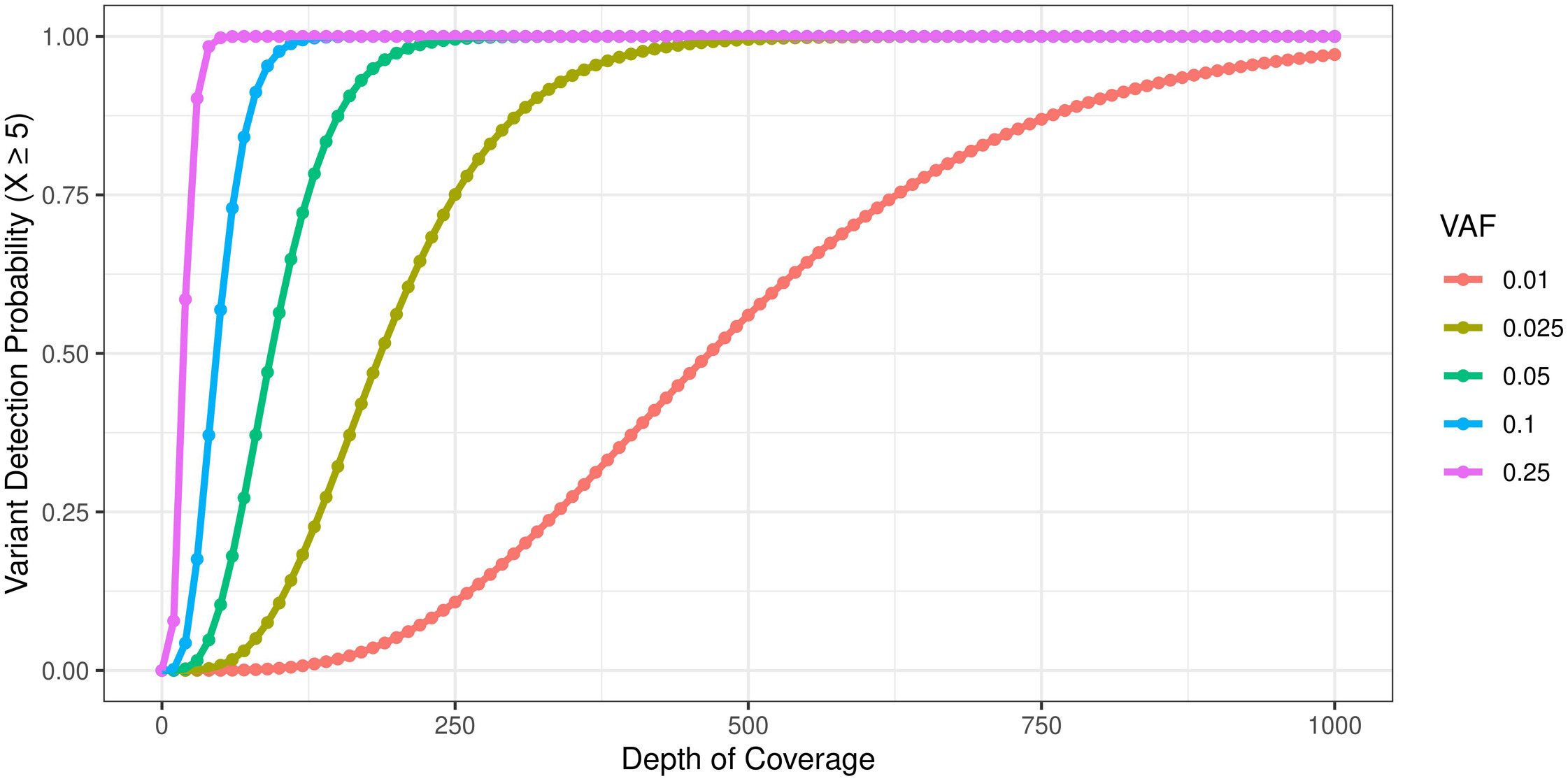
- Illustration of relationship between sequencing depth and variant call confidence as a function of VAF.
- Example of a valid VCF file with header and a few variant site records.
-
For hot spot testing, coverage of at least 100–300X is recommended.
-
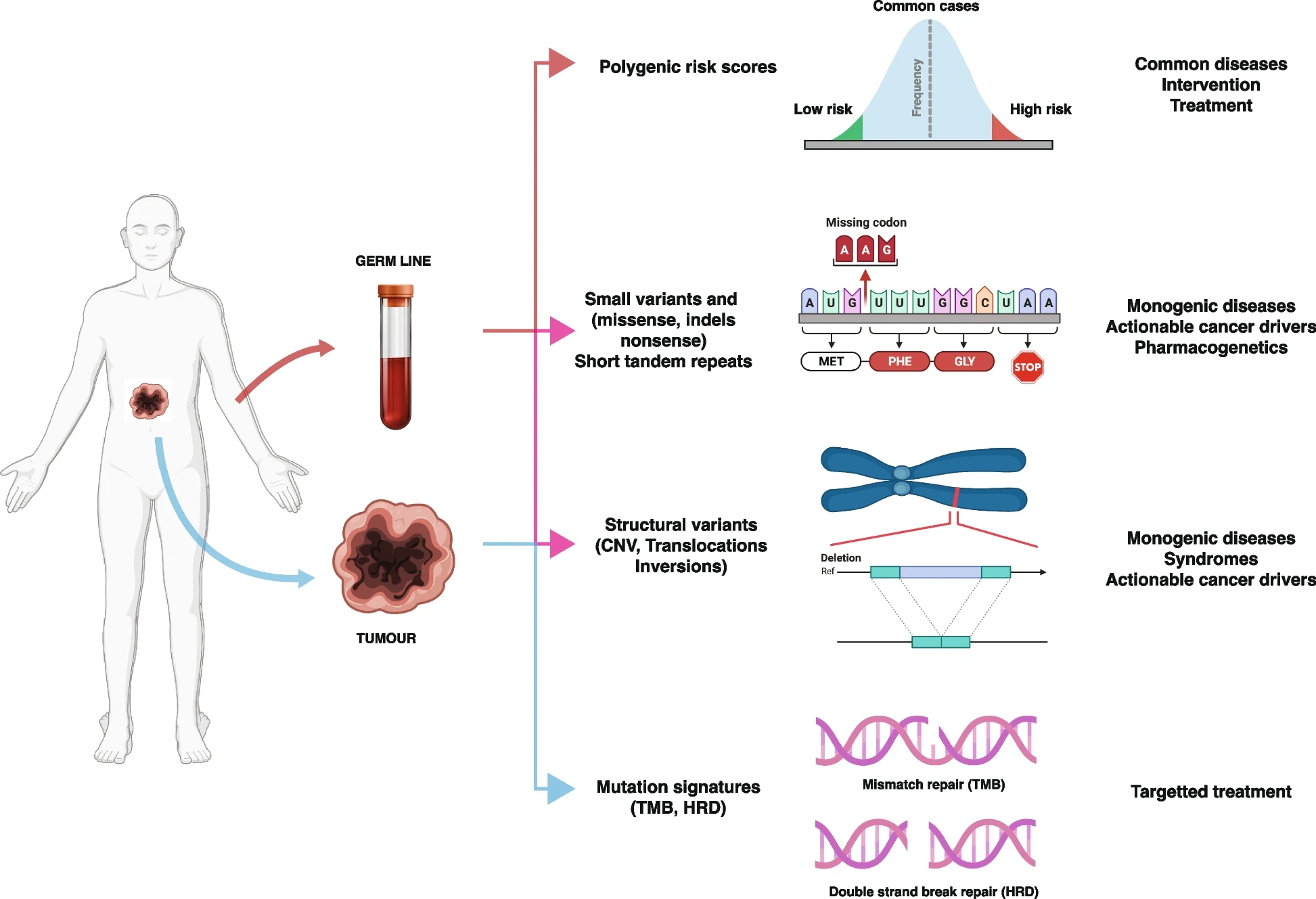
Whole Genome Sequencing (WGS) finds its primary clinical applications in diagnosing rare diseases and pinpointing actionable somatic variants within tumors.