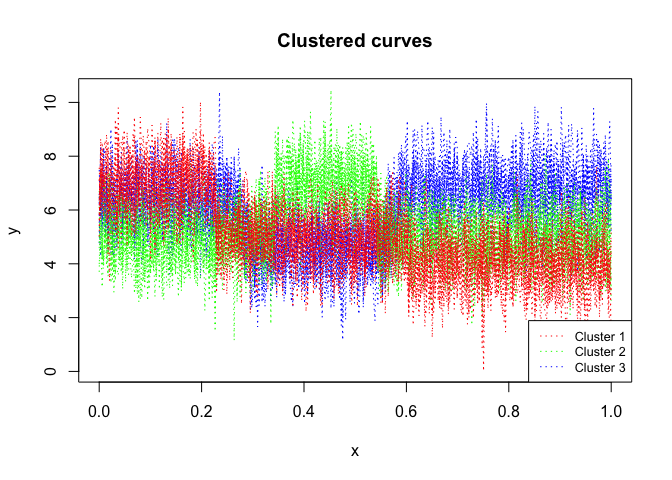
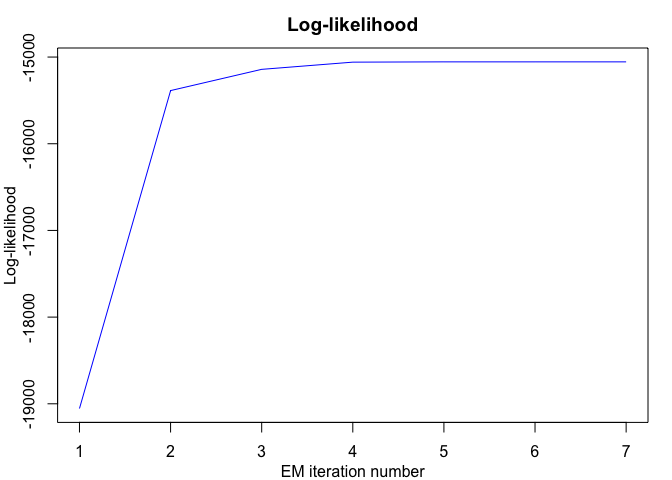
R code for the clustering and segmentation of time series (including with regime changes) by mixture of gaussian Hidden Markov Models (MixHMMs) and the EM algorithm, i.e functional data clustering and segmentation.
You can install the development version of mixHMM from GitHub with:
# install.packages("devtools")
devtools::install_github("fchamroukhi/mixHMM")To build vignettes for examples of usage, type the command below instead:
# install.packages("devtools")
devtools::install_github("fchamroukhi/mixHMM",
build_opts = c("--no-resave-data", "--no-manual"),
build_vignettes = TRUE)Use the following command to display vignettes:
browseVignettes("mixHMM")library(mixHMM)# Application to a toy data set
data("toydataset")
Y <- t(toydataset[,2:ncol(toydataset)])
K <- 3 # Number of clusters
R <- 3 # Number of regimes (HMM states)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
ordered_states <- TRUE
n_tries <- 1
max_iter <- 1000
init_kmeans <- TRUE
threshold <- 1e-6
verbose <- TRUE
mixhmm <- emMixHMM(Y = Y, K, R, variance_type,
ordered_states, init_kmeans, n_tries, max_iter, threshold,
verbose)
#> EM - mixHMMs: Iteration: 1 | log-likelihood: -19054.7157954833
#> EM - mixHMMs: Iteration: 2 | log-likelihood: -15386.7973253636
#> EM - mixHMMs: Iteration: 3 | log-likelihood: -15141.8435629464
#> EM - mixHMMs: Iteration: 4 | log-likelihood: -15058.7251666378
#> EM - mixHMMs: Iteration: 5 | log-likelihood: -15055.5058566489
#> EM - mixHMMs: Iteration: 6 | log-likelihood: -15055.4877310423
#> EM - mixHMMs: Iteration: 7 | log-likelihood: -15055.4876146553
mixhmm$summary()
#> -----------------------
#> Fitted mixHMM model
#> -----------------------
#>
#> MixHMM model with K = 3 clusters and R = 3 regimes:
#>
#> log-likelihood nu AIC BIC
#> -15055.49 41 -15096.49 -15125.21
#>
#> Clustering table (Number of curves in each clusters):
#>
#> 1 2 3
#> 10 10 10
#>
#> Mixing probabilities (cluster weights):
#> 1 2 3
#> 0.3333333 0.3333333 0.3333333
#>
#>
#> -------------------
#> Cluster 1 (k = 1):
#>
#> Means:
#>
#> r = 1 r = 2 r = 3
#> 7.00202 4.964273 3.979626
#>
#> Variances:
#>
#> Sigma2(r = 1) Sigma2(r = 2) Sigma2(r = 3)
#> 0.9858726 0.9884542 0.9651437
#>
#> -------------------
#> Cluster 2 (k = 2):
#>
#> Means:
#>
#> r = 1 r = 2 r = 3
#> 4.987066 6.963998 4.987279
#>
#> Variances:
#>
#> Sigma2(r = 1) Sigma2(r = 2) Sigma2(r = 3)
#> 0.9578459 1.045573 0.952294
#>
#> -------------------
#> Cluster 3 (k = 3):
#>
#> Means:
#>
#> r = 1 r = 2 r = 3
#> 6.319189 4.583954 6.722627
#>
#> Variances:
#>
#> Sigma2(r = 1) Sigma2(r = 2) Sigma2(r = 3)
#> 0.9571803 0.9504731 1.01553
mixhmm$plot()