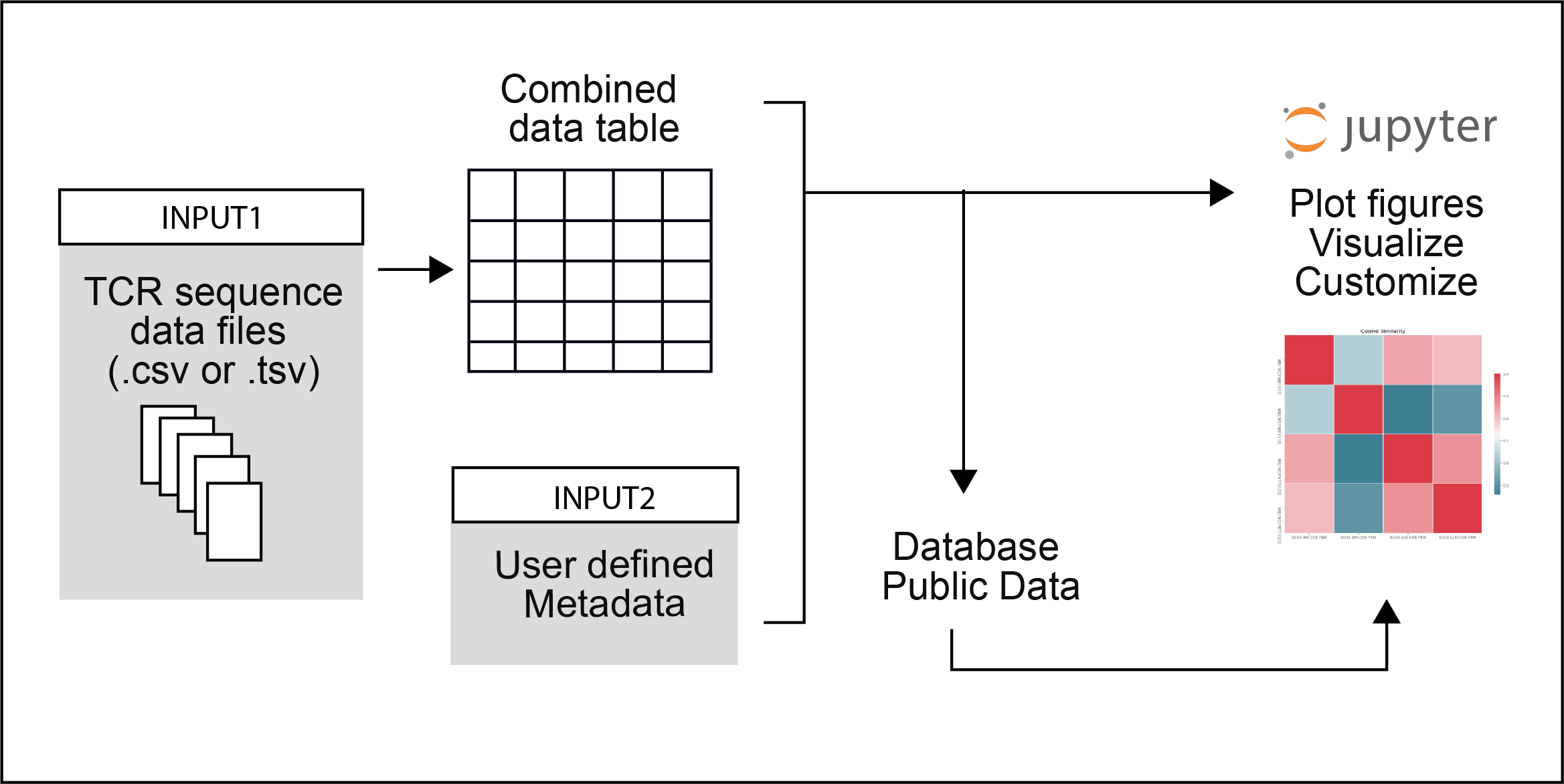
- Provide an interactive set of Jupyter notebooks for easily visualizing and analyzing TCR sequencing data using existing tools and methods.
- A program that inputs diverse file types from alignment softwares (ie. MIXCR, Changeo, and more) along with associated metadata for those files, and allows an analysis workflow.
-
T cells are immune cells that recognize their targets through the T-cell receptor (TCR) - a complex of highly variable cell-surface proteins. Analyzing the TCR repertoire in humans or mouse models can help us understand the development of the immune system and progression of disease.
-
There are a growing pool of biologists and clinicians that want to be able to analyze the mass amounts of data they are collecting, or that others have collected and published. These notebooks will provide a tool for this community to use and interact with T-cell receptor sequencing data.
-
There has been a lot of development of methods for analyzing T-cell receptor data, a lot of which borrows from the field of ecology and associated diversity analyses. However, the tools developed for these analyses are all in different locations and not easy to access! We are solving that problem here.
-
Notebooks have two input requirements:
Dependencies: pandas, jupyter, scipy, seaborn
-
You need python3 in order to install and use this. So first make sure you have python3.
-
clone
PyClonalrepo:$ git clone https://github.com/NCBI-Hackathons/PyClonal.git -
create virtual environment inside
PyClonaldirectory$ cd PyClonal $ python3 -m venv pyclonal -
activate virtualenv and install
PyClonal(this will install all necessary dependencies)$ source pyclonal/bin/activate $ pip install -e . -
open jupyter notebook within that environment inside
jupyter notebookdirectory$ cd jupyter_notebooks $ jupyter notebook -
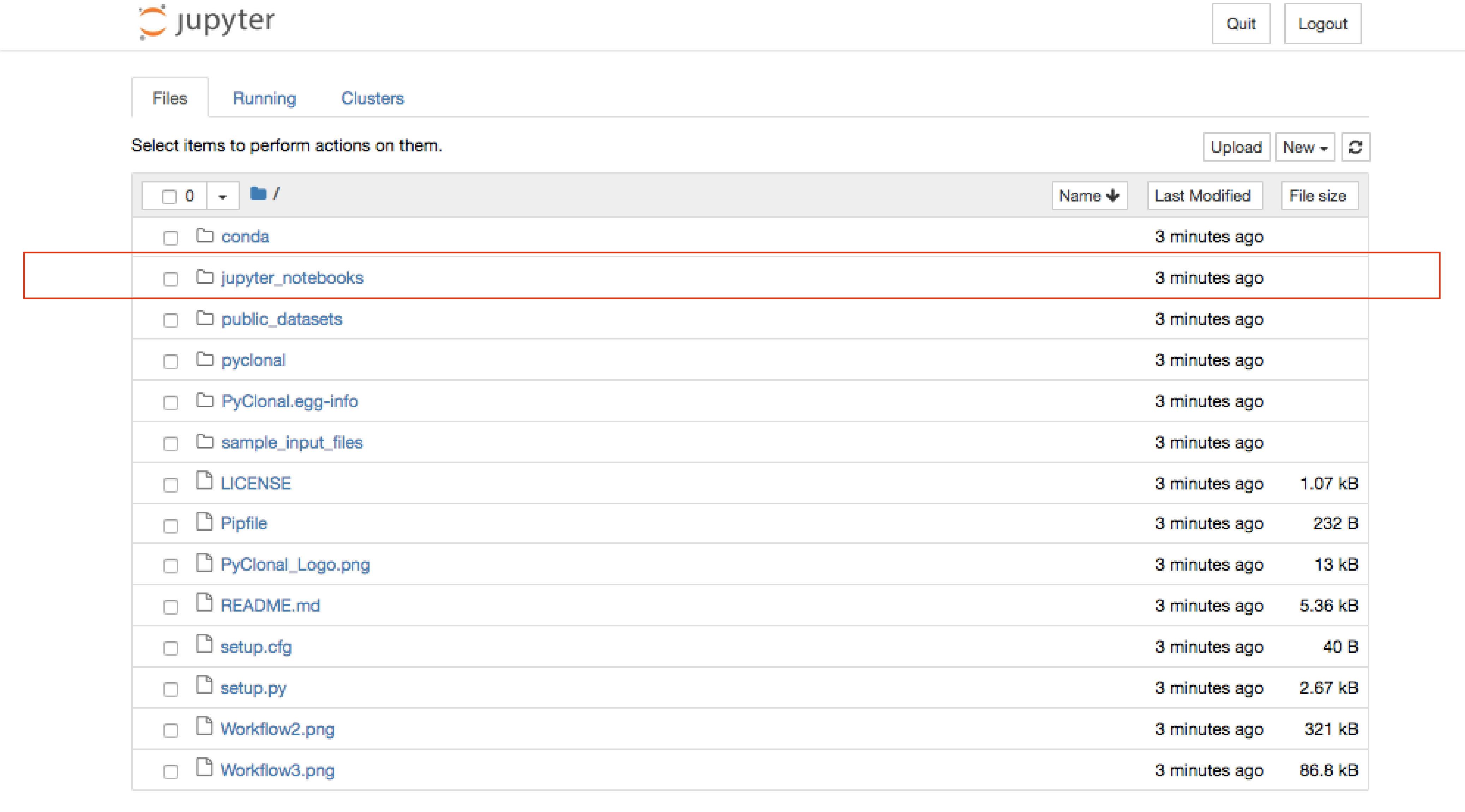
once the jupyter notebook browser launches as below, notebooks are in the jupyter_notebooks folder
-
to exit the environment type
exit -
to reopen the environment after it has been downloaded once
$ pipenv shell
Older versions of Anaconda have issues running pipenv. There are a few alternatives if you run into installation issues:
-
Update your version of Anaconda, and rerun the commands
-
Use a conda environment. In the PyClonal directory:
$ git clone https://github.com/NCBI-Hackathons/PyClonal.git $ cd PyClonal $ conda create --name env python=3 $ source activate env $ pip install -e . $ jupyter notebook -
Easiest but least recommended method:
$ git clone https://github.com/NCBI-Hackathons/PyClonal.git $ cd PyClonal $ pip install -e . $ jupyter notebook
To use from the command line, run pcl.py script:
$./pcl.py -h
usage: pcl.py [-h] [-p PATTERN] [-f [FORMAT [FORMAT ...]]] [-n FORMAT_NAME]
[-c [FORMAT_COLS [FORMAT_COLS ...]]] [-o OUTPUT_FILE]
dir
A Jupyter notebook based framework to analyze T-cell receptor sequencing data.
Provide an interactive set of Jupyter notebooks for easily visualizing and
analyzing TCR sequencing data using existing tools and methods.
positional arguments:
dir directory with data files
optional arguments:
-h, --help show this help message and exit
-p PATTERN, --pattern PATTERN
filename patterd (*.tsv)
-f [FORMAT [FORMAT ...]], --format [FORMAT [FORMAT ...]]
custom format: names of columns to extract
-n FORMAT_NAME, --format_name FORMAT_NAME
custom format name
-c [FORMAT_COLS [FORMAT_COLS ...]], --format_cols [FORMAT_COLS [FORMAT_COLS ...]]
column to detect format
-o OUTPUT_FILE, --output_file OUTPUT_FILE
output files basename
For usage example in jupyter notebook see example notebook data input.ipynb
in jupyter_notebooks directory.
-VDJdb -https://vdjdb.cdr3.net/
-VDJ tools -https://vdjtools-doc.readthedocs.io/en/master/
-VDJviz: a versatile immune repertoire browser -https://vdjviz.cdr3.net/
-tcR -https://cran.r-project.org/web/packages/tcR/vignettes/tcrvignette.html
-miXCR -https://mixcr.readthedocs.io/en/master/
-powerTCR -https://www.biorxiv.org/content/early/2018/04/07/297119
-ImmuneDB -http://immunedb.com/
-TraCeR -https://github.com/teichlab/tracer