Notice: The GUI is currently not functionally because of some underlying changes and outdated dependencies. We hope to fix this in the upcoming weeks. Feel free to use the toolkit in the meantime to access the same functionality through a python API.
NeuroX is a comprehensive toolkit based on explainable AI research published in AAAI'19 [1] and ICLR'19 [2]. It facilitates the analysis of individual neurons in deep neural networks. Currently support is provided for sequence-to-sequence models trained using a slightly modified version of the OpenNMT-py toolkit [3]. Support for other toolkits and architectures like Transformers is planned for the near future.
The toolkit allows the user to upload a model and rank neurons based on their saliency to the overall model, or to some specific property that is being learned intrinsically learned inside a deep neural network (e.g. Part of Speech tagging inside a machine translation model, position detector inside a language model etc.)
The toolkit is written using Python 3 and is accessible through a browser as a web application. Extensive testing has been doing using Google Chrome, but the toolkit should be functional in other browsers as well. The toolkit is meant to be used as Jupyter Notebooks are used - a server is launched (either on your local machine or on a more powerful server), and the application is then accessible using a URL.
If you wish to just try out the toolkit with a pre-trained model, a docker image is available with all of the required dependencies. Run the following to automatically download and run the server, after which you can follow steps 5-8 under Running the Server:
docker run -p 5001:5001 --rm fdalvi/neurox:v1- Conda installation (Ananconda/Miniconda)
- Python 3
- Node v10
- Getting the code:
git clone https://github.com/fdalvi/NeuroX.git
cd neuroX/
git submodule init
git submodule update- Setting up the environment:
conda env create -f conda-environment.yml -n neurox-env
source activate neurox-env- Building the frontend:
cd src
npm install
npm run build- Running the server:
python app.pyThe server will be up and running on the localhost at port 5001. You can set
any environment variables such as CUDA_VISBLE_DEVICES or CUDA_CACHE_PATH and
they will be used for the computation that will be done later.
-
Accessing the toolkit: Visit
http://localhost:5001orhttp://<server-ip>:5001 -
(a) Starting a new project:
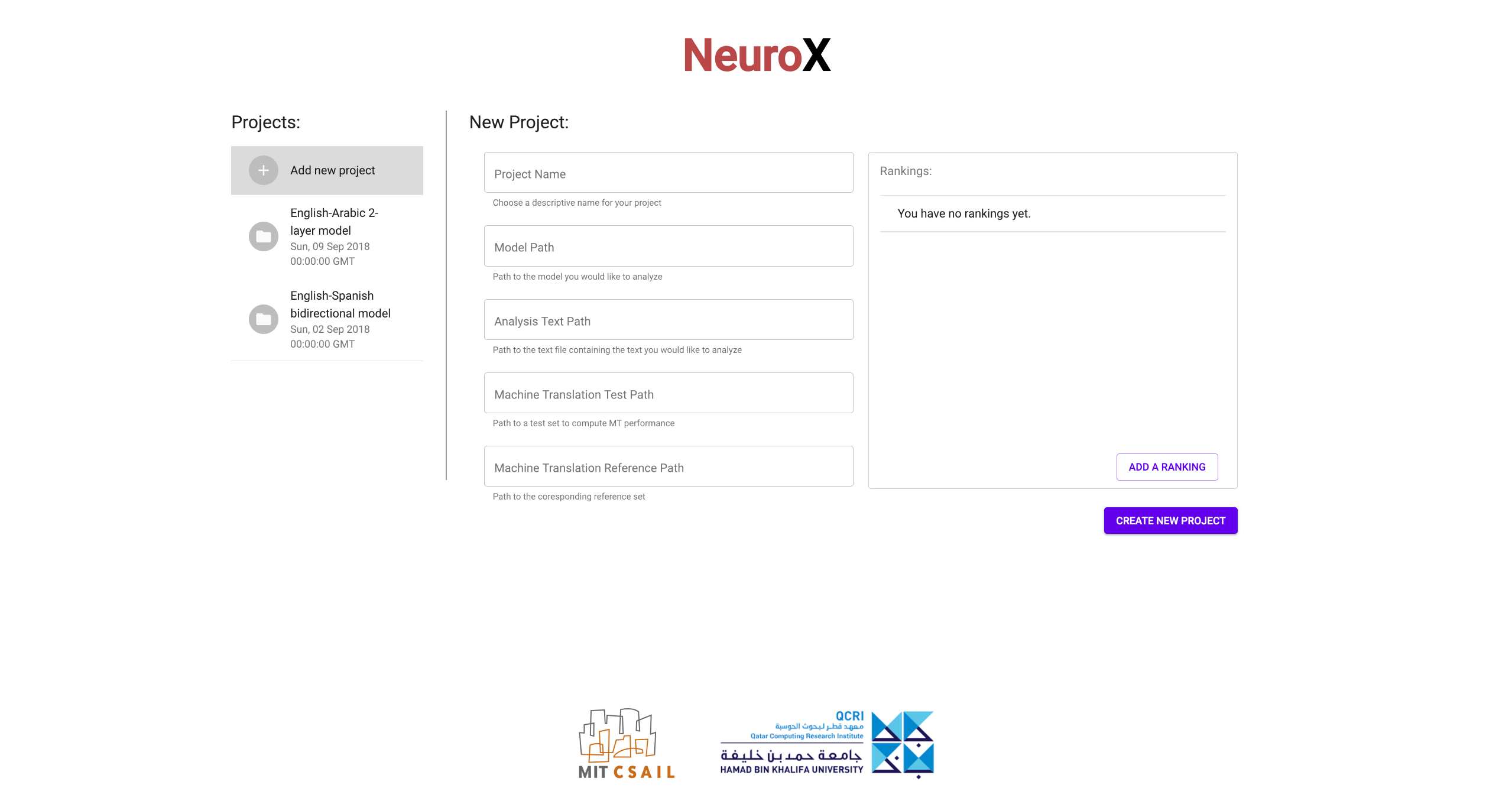 Fill in the required details to start a new project. You will need to provide
the path to a OpenNMT-py model trained using
[3]. You will also need to
provide the path to a text file which will serve as the input to your model for
analysis. Finally, you will also need to provide a ground truth test set
(input and output reference) to run the ablation studies. Apart from this basic
information, you can add an arbitrary number of ranking schemes using the
rankings dialog on the right (See section
Rankings Overview
for an explanation of the various ranking schemes).
Fill in the required details to start a new project. You will need to provide
the path to a OpenNMT-py model trained using
[3]. You will also need to
provide the path to a text file which will serve as the input to your model for
analysis. Finally, you will also need to provide a ground truth test set
(input and output reference) to run the ablation studies. Apart from this basic
information, you can add an arbitrary number of ranking schemes using the
rankings dialog on the right (See section
Rankings Overview
for an explanation of the various ranking schemes). -
(b) Loading an existing project: Just select the project from the sidebar to see it's details and load an existing project.
-
Waiting for Computation: Depending on the number of rankings and the size of your models/test data, it will take a while to compute all of the information required to analyze the neurons. You will see a loading screen while this is taking place:
 It is highly recommended to run the server with access to a GPU to speed up the
computation. The analysis requires running the trained OpenNMT-py model several
times to extract activations, train several classifiers to generate the
rankings, run several evaluations to compute the ablation results and also run
the model in real-time to manipulate the results. All of these tasks are
significantly faster on a GPU. All of the computation for a project is done
once and cached, and hence you can leave the page and come back to it later and
all of the results will be available instantaneously.
It is highly recommended to run the server with access to a GPU to speed up the
computation. The analysis requires running the trained OpenNMT-py model several
times to extract activations, train several classifiers to generate the
rankings, run several evaluations to compute the ablation results and also run
the model in real-time to manipulate the results. All of these tasks are
significantly faster on a GPU. All of the computation for a project is done
once and cached, and hence you can leave the page and come back to it later and
all of the results will be available instantaneously. -
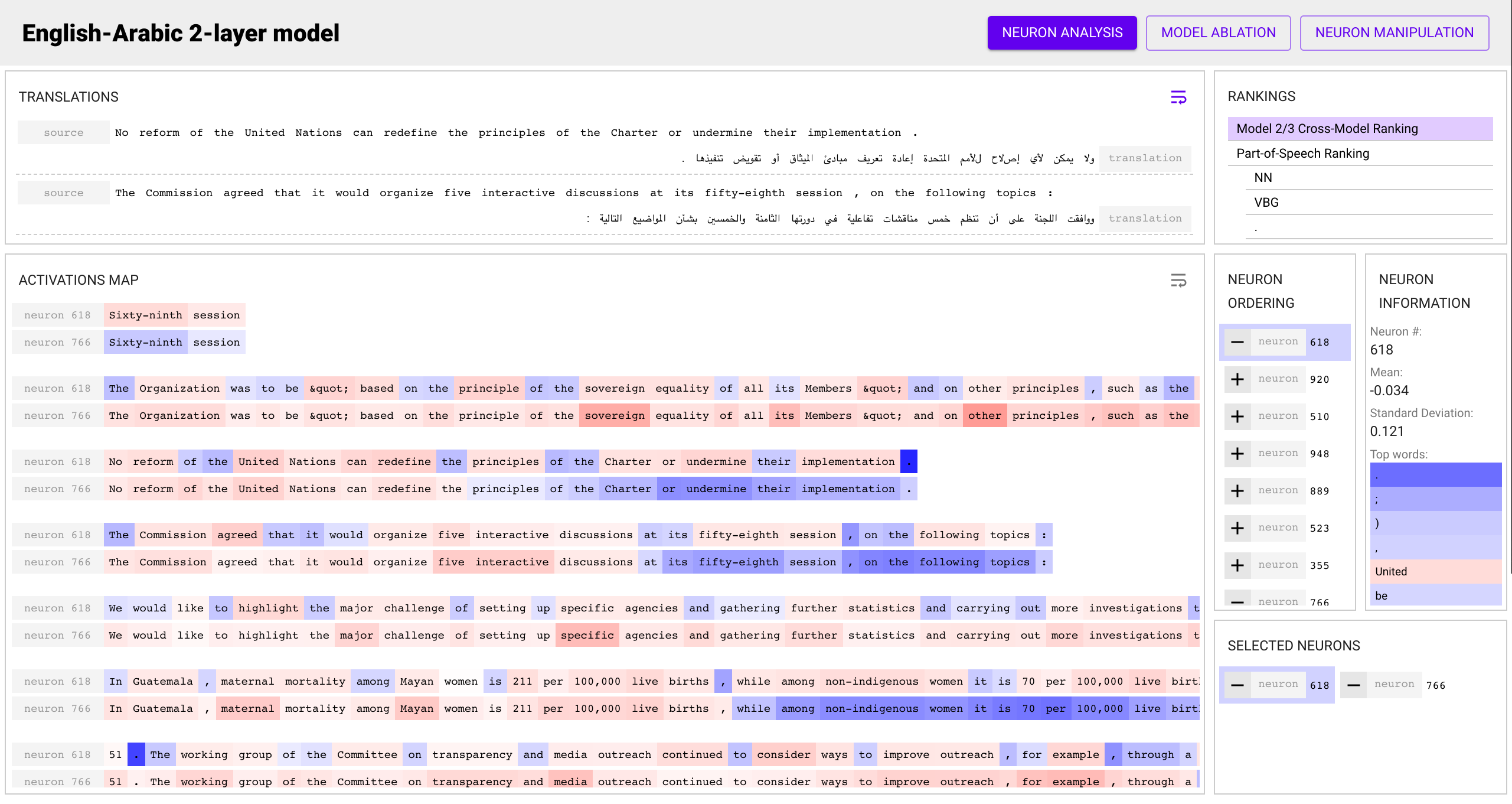
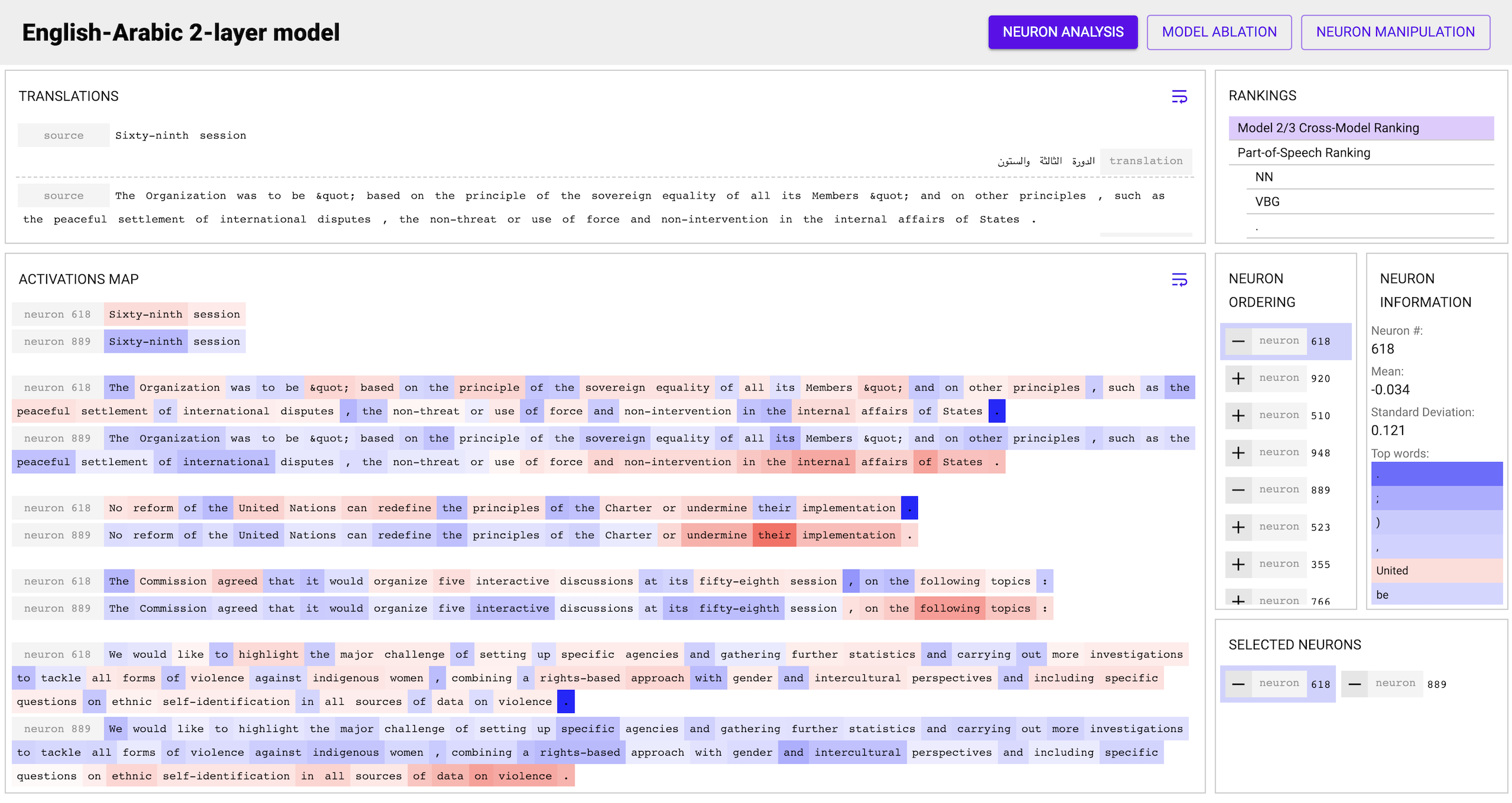
Perform your analysis! The toolkit provides three major views:
- The Analysis view allows you to view the various rankings, select
multiple neurons to see their activation maps on the input data, and look
at statistics for individual neurons such as their top words.
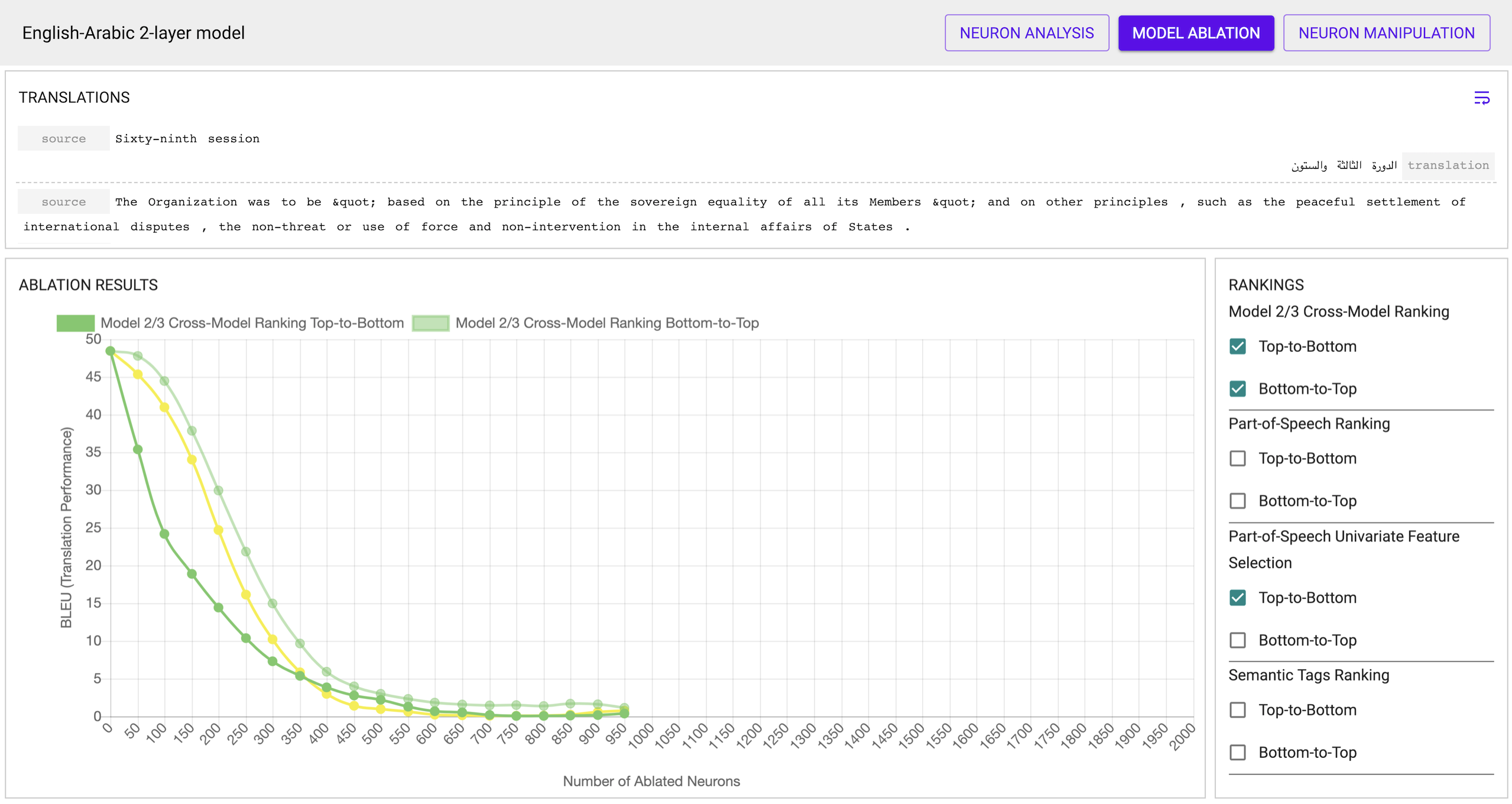
- The Ablation view allows you to view the results of ablation using the
various rankings. This information can be useful to (i) verify the
correctness of the rankings, and (ii) hypothesis what information is more or
less important to the network
- The Manipulation view allows you to control the activation patterns of
individual neurons and see the effect on the target in real-time
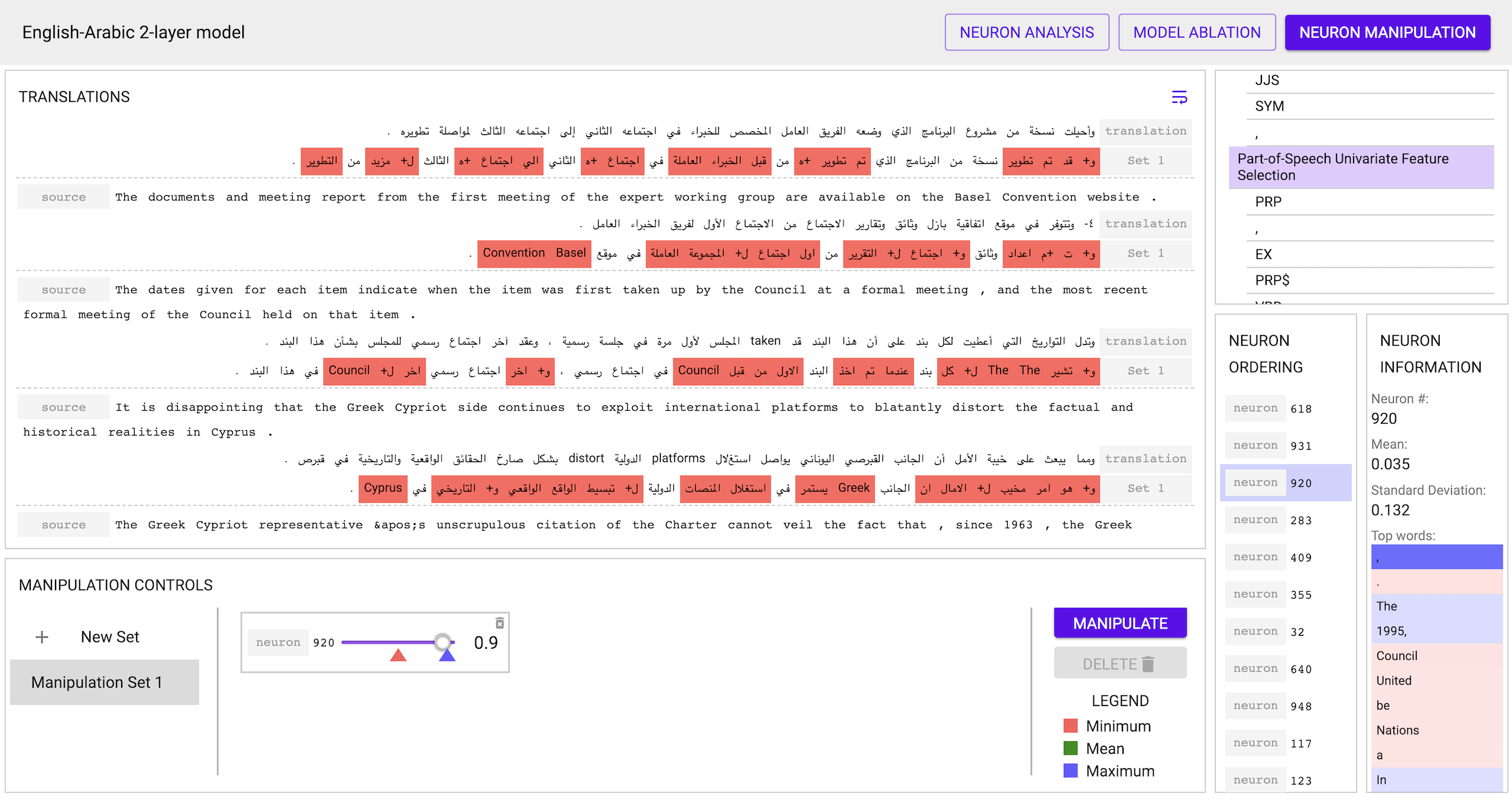
- The Analysis view allows you to view the various rankings, select
multiple neurons to see their activation maps on the input data, and look
at statistics for individual neurons such as their top words.
Various ranking schemes can be used to rank the individual neurons in a given model:
- Cross-Model Correlation: Rank neurons by how correlated their activation patterns are to neurons in other networks trained to perform the same task. This method is unsupervised and just requires two or more models trained for the same task.
- Linguistic Correlation: Rank neurons by how well they predict a given property. This method is supervised, and requires a annotated set where each input word is tagged with a property of interest
- Univariate Feature Selection: Similar to Linguistic Correlation, but applies a different criteria to rank the neurons.
Please see the links below for details behind each method and how the rankings are actually computed.
- [1] AAAI'19 Publication: What Is One Grain of Sand in the Desert? Analyzing Individual Neurons in Deep NLP Models
- [2] ICLR'19 Publication: Identifying And Controlling Important Neurons In Neural Machine Translation
- [3] Modified OpenNMT-py toolkit to train the models that are to be analyzed
- [4] AAAI'19 Demonstration: NeuroX: A Toolkit for Analyzing Individual Neurons in Neural Networks
- Restructuring for better modularity
- DB for backend
- Client side caching for requests
- Computation queue on the backend
- Interaction with classifier/correlation/translation modules
- Manipulation UI
- Real time Logs for computation
- Ranking confidence measures
- BPE/Char/Segmented support
- LM support
- General model support
- Support for file browser to select models/test files