This project is about using driver CUDA to execute the code faster on the GPU than on the CPU.
I simulated the Ising model (a simple model for ferromagnetism in matter) employing a MonteCarlo Mark Chain (MCMC). The simulation is performed in C (using CUDA driver), the analysis results are computed in python.
A time comparison between pure C and Cuda is reported below.
-
Windows 10 or a Linux distribution with a compatible kernel (https://docs.nvidia.com/cuda/cuda-installation-guide-linux/index.html#system-requirements)
-
A compatible GPU (see https://developer.nvidia.com/cuda-gpus)
-
Python 3
-
Clone this repo on your computer
$ git clone https://github.com/federicovisintini/ising-gpu.git
-
(recommended) Create a virtual enviroment
$ cd ising-gpu $ python3 -m venv isingvenvand activate it
$ source isingvenv/bin/activate -
Install the requirements
$ pip install -r requirements.txt
-
Install CUDA following the offical docs
The C and the CUDA files will generate a Markov Chain of energy and magnetisation of the system each.
Results are saved in /data in .dat files.
-
Run C code with the following
-
Modify the relevant parameters in lines 7-13 of
dim2.c, expecially the path where the results will be saved in and the lattice spacing -
Compile
dim2.cin an executable calleddim2cwith$ gcc dim2.c -o dim2c -lm
-
Execute
dim2cto generate result$ ./dim2c
Note: to time the execution time just type
$ time ./dim2cinstead.
-
-
Run CUDA code with the following
-
Modify the relevant parameters in lines 11-19 of
dim2.cu, expecially the path where the results will be saved in and the lattice spacing -
Compile
dim2.cuin an executable calleddim2cuwith$ nvcc dim2.cu -o dim2cu -lcurand
-
Execute
dim2cuto generate result$ ./dim2cu
Note: to time the execution time just type
$ time ./dim2cuinstead.
-
-
Error analysis at given lattice spacing and temperature
-
Run
$ python single_temperature_err_analysis.pyto compute the mean quantities (energy, magnetization, suscettibility) and associated errors. techniques are used to take into account autocorrelation between the measures.The script runs on files named
data/lattice*cu.dat.
-
-
Run
$ python multiple_temperature.pyto a comprehesive analysis of mean quantities around phase transition temperature and dependance from latting spacing.The script runs on files named
data/lattice*cu.dat.
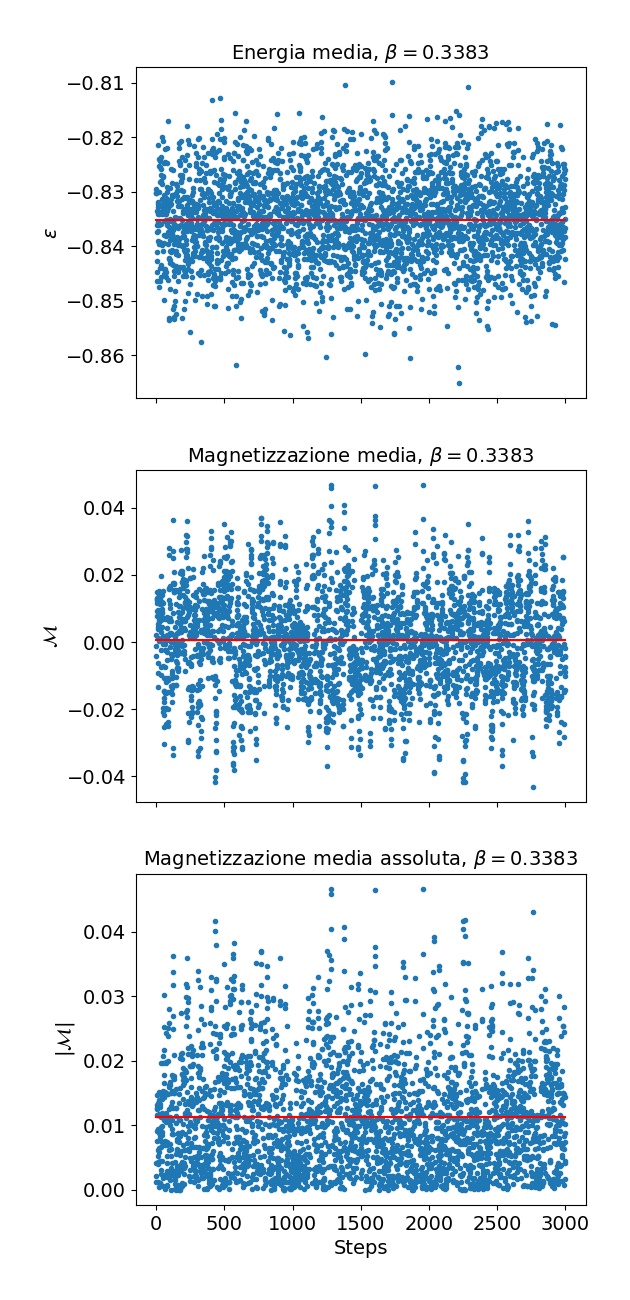
Plotting the energy and magnetization vs its position in the markov chain should result in something like this:
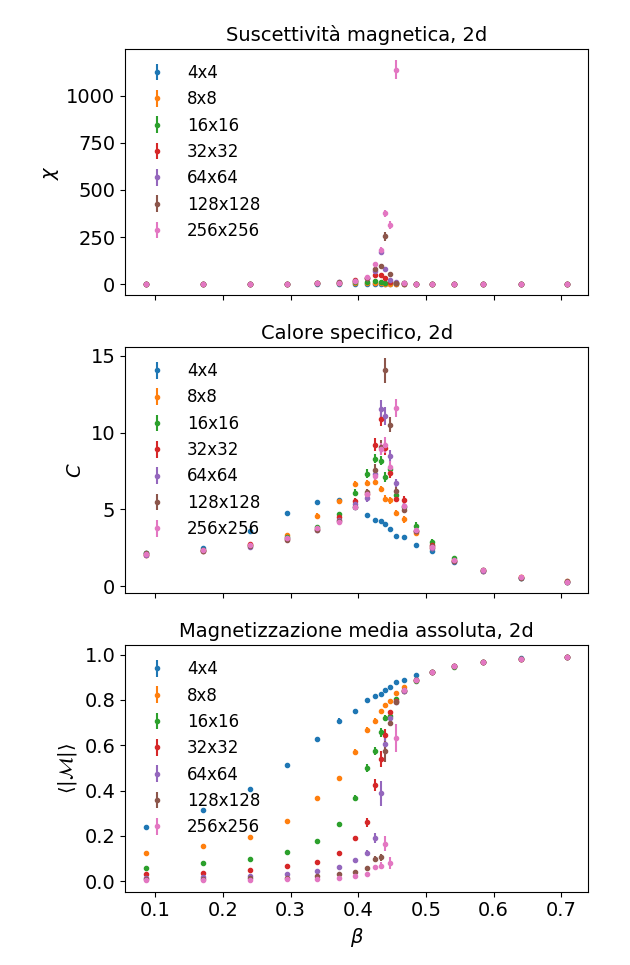
Computing suscettibility, specific heat and magnetization varying the temperature (β~1/T) should result instead in something like this:
We compared the time it took to execute a simulation of our system for some fix parameter and varying the lattice dimension for C and CUDA scrips.
The number of steps between measures is proportional to lattice x lattice, so we expect a quadratic (in 2D) increase in computational time (x4 in table rows, since doubling the lattice points each time).
| lattice | C code runtime (s) | CUDA code runtime (s) |
|---|---|---|
| 4 | 0.221 | 5.364 |
| 8 | 0.719 | 5.597 |
| 16 | 2.431 | 5.723 |
| 32 | 9.806 | 7.564 |
| 64 | 38.655 | 14.874 |
| 128 | 148.755 | 47.485 |
| 256 | 630.897 | 169.416 |
We see that for a relatively large lattice (ca >30 points), the CUDA program is actually faster than the C code.
This work is licenced under GNU GPLv3 by Federico Visintini.
See LICENSE for more information.