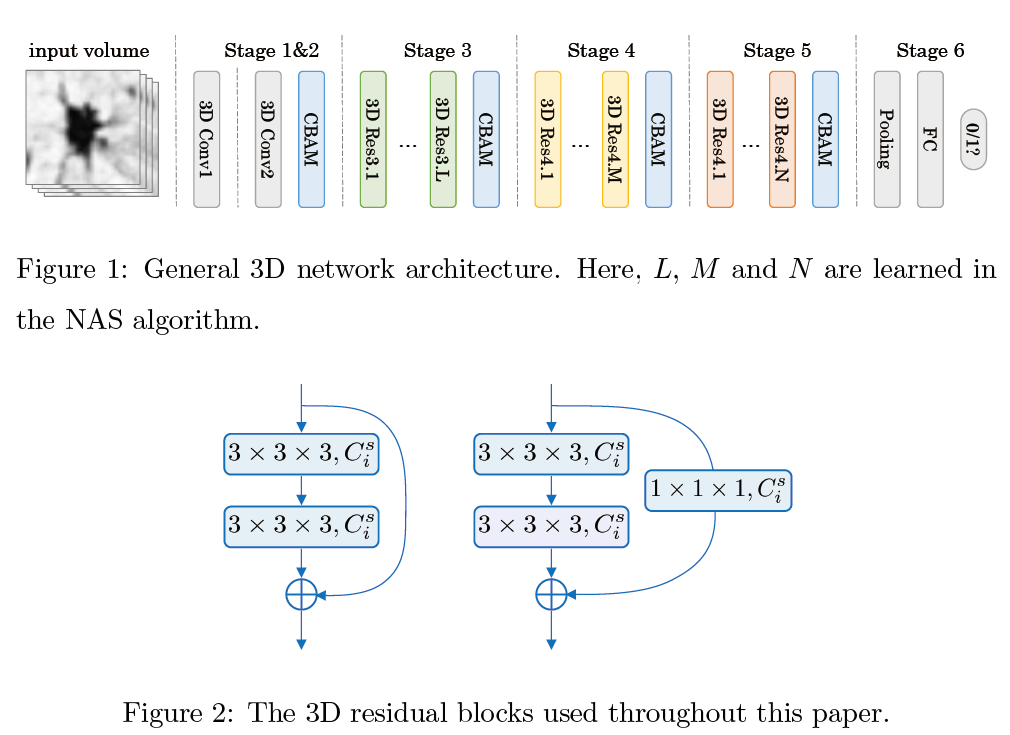
3D Neural Architecture Search (NAS) for Pulmonary Nodules Classification
Hanliang Jiang, Fuhao Shen, Fei Gao*, Weidong Han. Learning Efficient, Explainable and Discriminative Representations for Pulmonary Nodules Classification. Pattern Recognition, 113: 107825, 2021.
@article{Jiang2021naslung, author = {Hanliang Jiang and Fuhao Shen and Fei Gao and Weidong Han}, title = {Learning efficient, explainable and discriminative representations for pulmonary nodules classification}, journal = {Pattern Recognition}, volume = {113}, pages = {107825}, year = {2021}, issn = {0031-3203}, doi = {https://doi.org/10.1016/j.patcog.2021.107825}, }
| model | Accu. | Sens. | Spec. | F1 Score | para.(M) |
|---|---|---|---|---|---|
| Multi-crop CNN | 87.14 | - | - | - | - |
| Nodule-level 2D CNN | 87.30 | 88.50 | 86.00 | 87.23 | - |
| Vanilla 3D CNN | 87.40 | 89.40 | 85.20 | 87.25 | - |
| DeepLung | 90.44 | 81.42 | - | - | 141.57 |
| AE-DPN | 90.24 | 92.04 | 88.94 | 90.45 | 678.69 |
| NASLung (ours) | 90.77 | 85.37 | 95.04 | 89.29 | 16.84 |
| Model | Accu. | Sens. | Spec. | F1 Score | para. |
|---|---|---|---|---|---|
| Model-1 | 88.83 | 87.20 | 90.12 | 87.50 | 0.14 |
| Model-2 | 88.42 | 84.38 | 91.46 | 86.67 | 2.61 |
| Model-3 | 88.17 | 84.44 | 91.60 | 86.50 | 3.90 |
| Model-4 | 88.13 | 83.20 | 92.28 | 86.30 | 2.54 |
| Model-5 | 87.97 | 83.72 | 91.31 | 86.22 | 0.43 |
| Model-6 | 87.77 | 83.67 | 91.00 | 86.03 | 0.22 |
| Model-7 | 87.76 | 84.14 | 89.79 | 85.98 | 0.86 |
| Model-8 | 88.00 | 82.43 | 92.69 | 85.97 | 4.02 |
| Model-9 | 88.04 | 78.01 | 96.09 | 85.36 | 4.06 |
| Model-10 | 87.22 | 82.70 | 90.92 | 85.32 | 0.24 |
- Linux or similar environment
- Python 3.7
- Pytorch 0.4.1
- NVIDIA GPU + CUDA CuDNN
-
Clone this repo:
git clone https://github.com/fei-hdu/NAS-Lung cd NAS-Lung -
Install PyTorch 0.4+ and torchvision from Pytorch and other dependencies (e.g., visdom and dominate). You can install all the dependencies by
pip install -r requirments.txt
-
Download Dataset LIDC-IDRI
python search_main.py --train_data_path {train_data_path} --test_data_path {test_data_path} --save_module_path {save_module_path}-
Train a model
sh run_training.sh
-
Test a model
python test.py --test_data_path {test_data_path} --preprocess_path {preprocess_path} --model_path {model_path}
- our final result can be download:Google Drive
- Best practice for training and testing your models.
- Feel free to ask any questions about coding. Fuhao Shen,
1048532267sfh@gmail.com
- Our work/code is inspired by Partial Order Pruning: for Best Speed/Accuracy Trade-off in Neural Architecture Search, CVPR 2019.
- S. Armato III, G. et al., Data from LIDC-IDRI, The Cancer Imaging . LIDC-IDRI.
- X. Li, Y. Zhou, Z. Pan, J. Feng, Partial order pruning: For best speed/accuracy trade-off in neural architecture search (2019) 9145–9153.
- S. Woo, J. Park, J.-Y. Lee, I. So Kweon, CBAM: Convolutional block attention module, in: Proceedings of the European Conference on Computer Vision (ECCV), 2018, pp. 3–19.
- W. Liu, Y. Wen, Z. Yu, M. Li, B. Raj, L. Song, Sphereface: Deep hypersphere embedding for face recognition, in: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017.
- T. Elsken, J. H. Metzen, F. Hutter, Neural architecture search: A survey, Journal of Machine Learning Research 20 (55) (2019) 1–21.
- W. Zhu, C. Liu, W. Fan, X. Xie, Deeplung: Deep 3d dual path nets for automated pulmonary nodule detection and classification, in: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), IEEE, 2018, pp. 673–681.