This repository contains two implementations of the Ant Colony Optimization algorithm
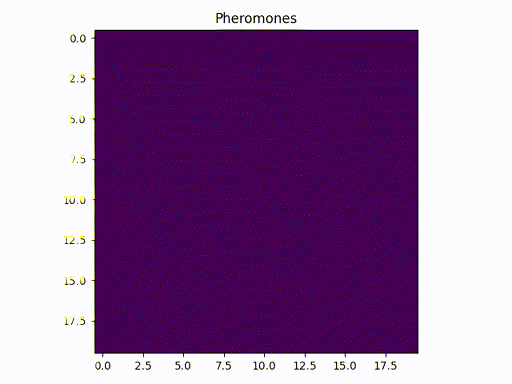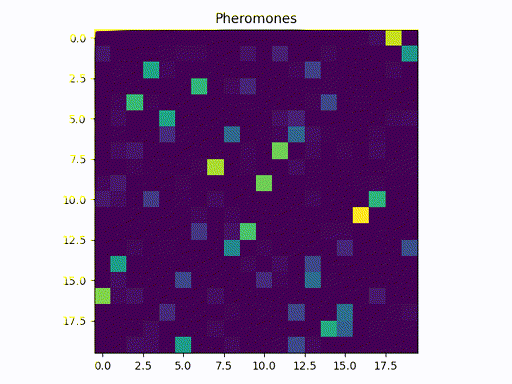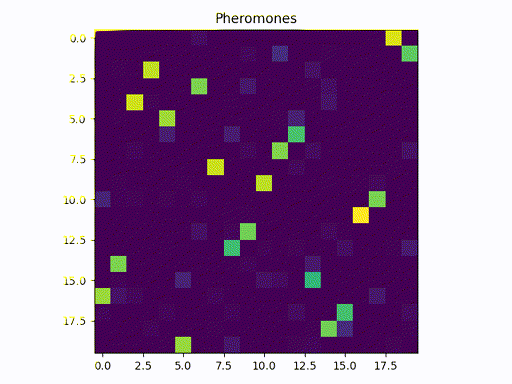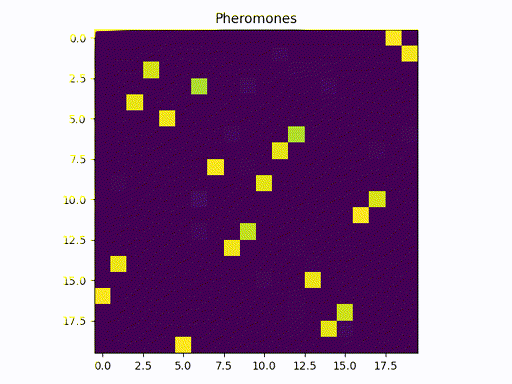
This image represents a graph adjacency matrix. We can cleary see that the pheromone concentrates in a specific path (one square per line)
Install dependencies: pip3 install -r python_implementation/requirements.txt --user
Run: python3 python_implementation/main.py
Usage
python3 python_implementation/main.py -h
usage: Ant Colony Optimization - Longest Path [-h] [-i INPUT] [-p POP_SIZE] [-a ALPHA] [-b BETA] [-e EVAPORATION] [-m MAX_ITERATIONS]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
Input graph
-p POP_SIZE, --pop-size POP_SIZE
Number of ants
-a ALPHA, --alpha ALPHA
Pheromone weight
-b BETA, --beta BETA Desirability weight
-e EVAPORATION, --evaporation EVAPORATION
Pheromone evaporation
-m MAX_ITERATIONS, --max-iterations MAX_ITERATIONS
Max Iterations
Install dependencies: CUDA
Compile: make
Run:
mkdir -p results/./ACO_red bases_grafos/entrada1.txt 100 50 0.2 1 2 test_exp
Usage
./ACO <input database> <N_ITER> <N_ANTS> <EVAPORATION RATE> <ALPHA> <BETA> [exp_name]
Positional Arguments:
input database - Input graph
N_ITER - Max Iterations
N_ANTS - Number of ants
EVAPORATION RATE - Pheromone evaporation
ALPHA - Pheromone weight
BETA - Desirability weight
exp_name - [OPTIONAL] Save results in file results/[exp_name].txt