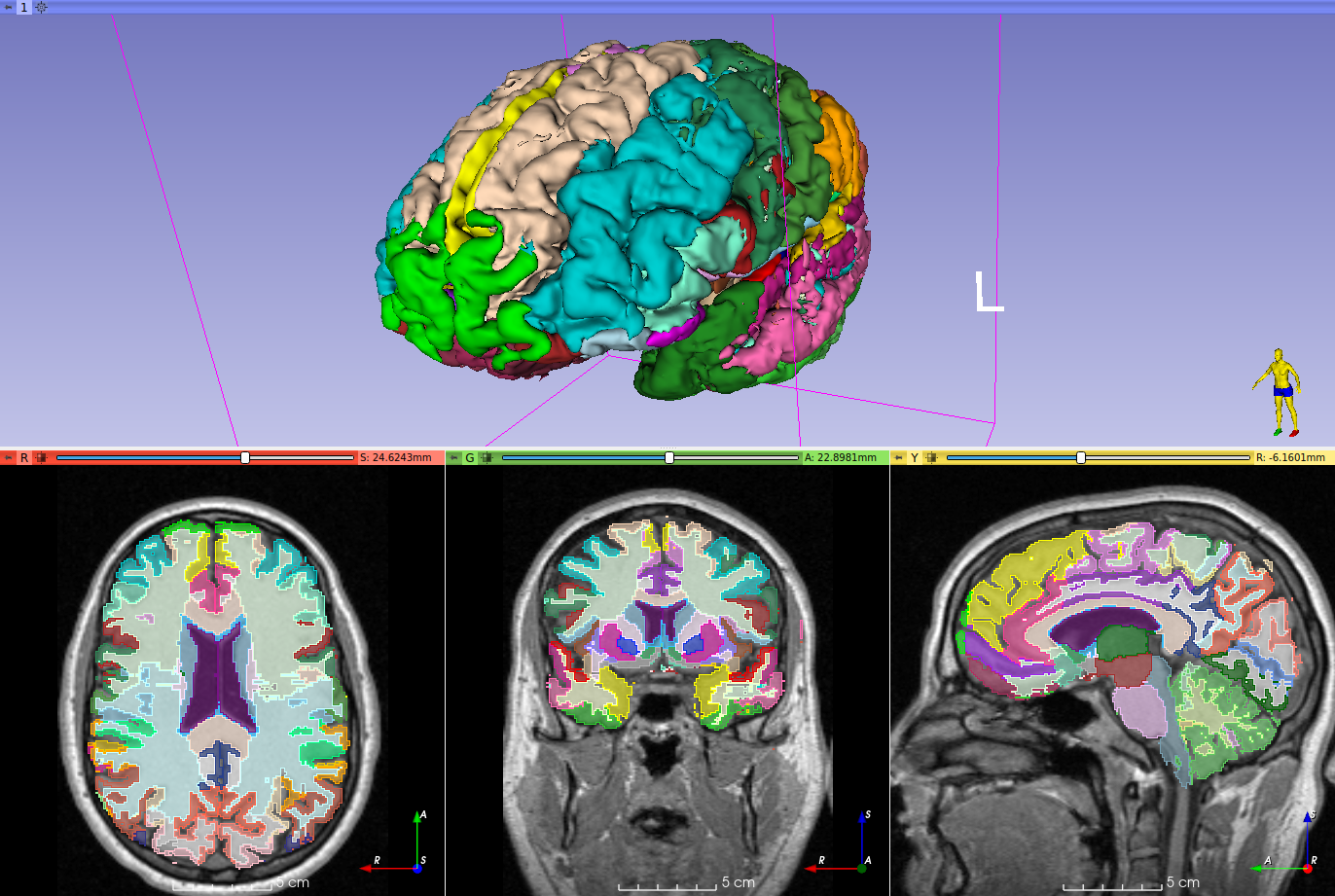
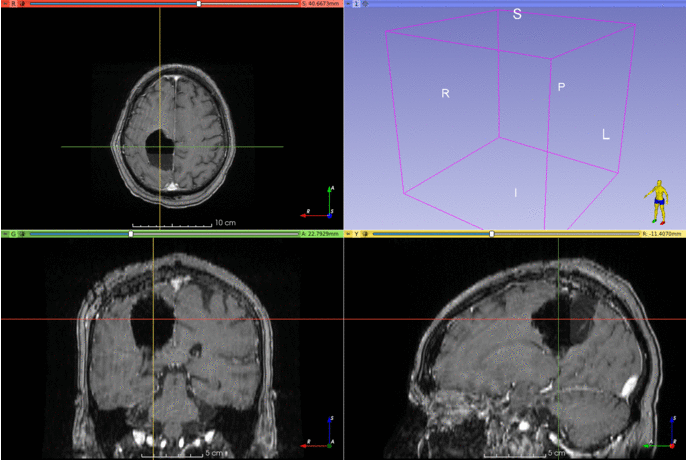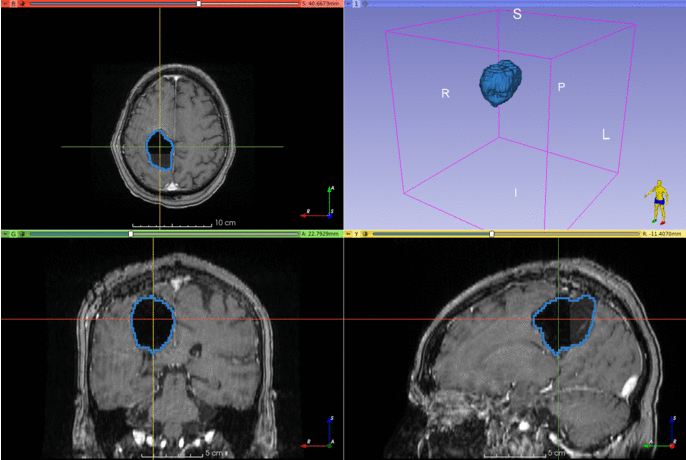
This repository contains the code for two Slicer modules that can be used to segment brain structures on T1-weighted MRIs.
Segmentations are performed using convolutional neural networks (CNNs), i.e., deep learning models. They take less than a minute on a graphics processing unit (GPU).
This is a project for the 35th NA-MIC Project Week.
git clone https://github.com/fepegar/SlicerParcellation.gitDownload the zipped directory and unzip it.
In Slicer, go to Edit -> Application Settings -> Modules and add the cloned/downloaded folder to the Additional module paths. When prompted, restart Slicer.
It splits the brain in 160 different structures, similar to the geodesic information flows (GIF) algorithm by Cardoso et al. 2015.
The PyTorch model was ported from NiftyNet at the MICCAI Educational Challenge 2019: Combining the power of PyTorch and NiftyNet.
The highresnet Python package can be installed running pip install highresnet to parcellate images outside 3D Slicer.
The segmentation works best if the input images are in the MNI space. The model was trained on T1-weighted MRIs with simulated resections, but it seems to work well with T1 images with gadolinium as well.
Sample images from the EPISURG dataset can be used to try this module.
The resseg Python package can be installed to segment cavities outside 3D Slicer.