Brain vessel segmentation from digital subtraction angiography (DSA) using a 3D convolutional neural network (CNN).
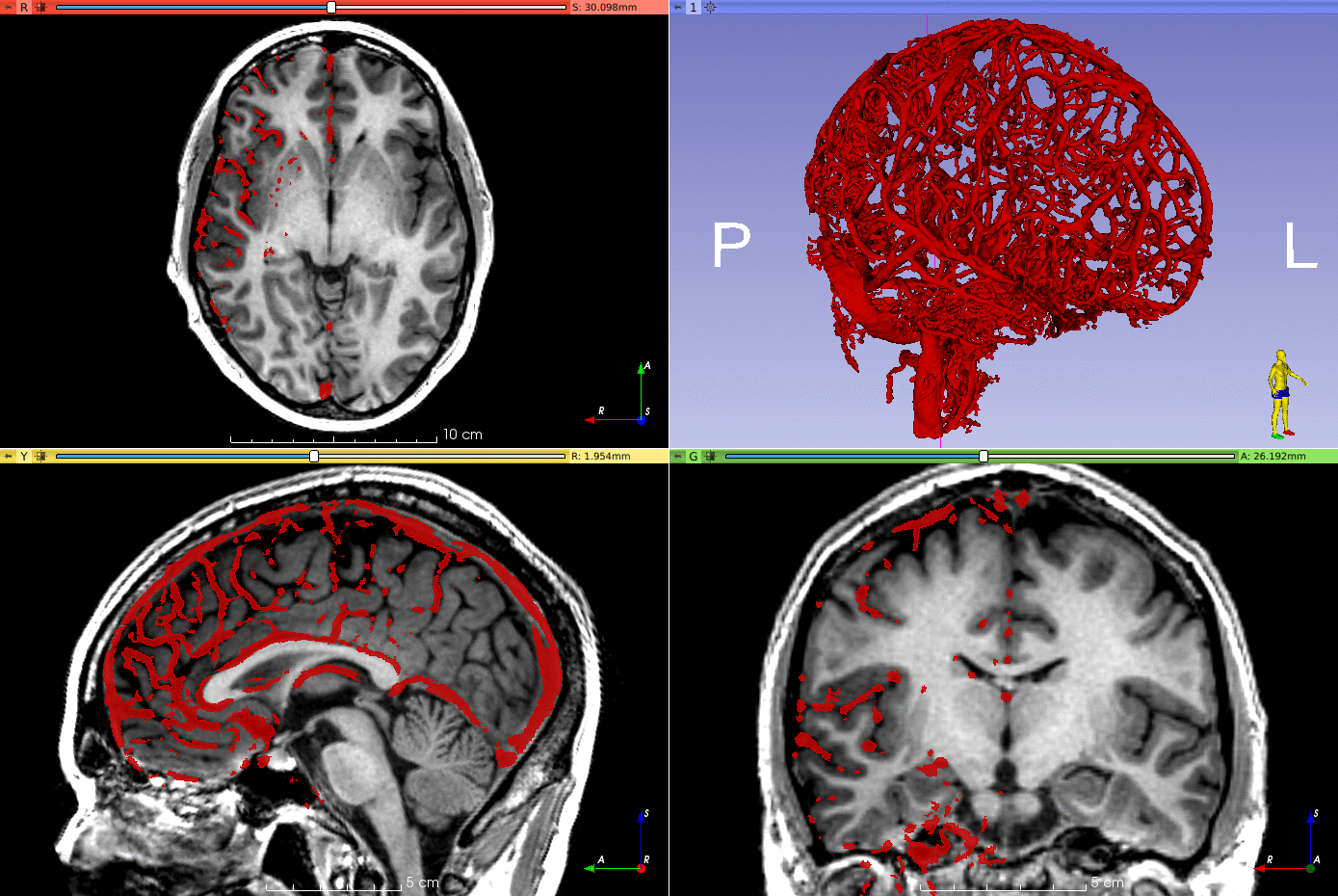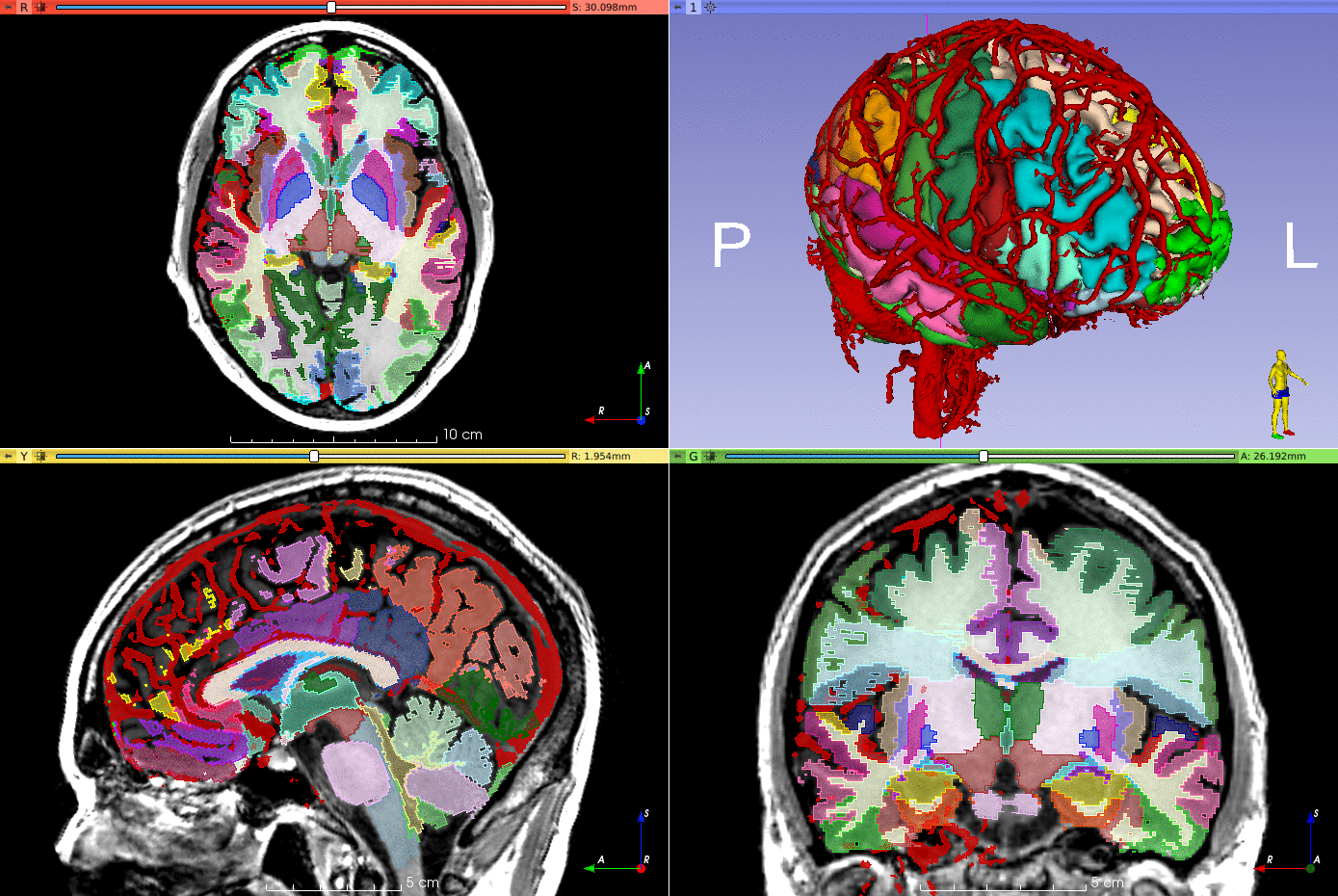 (Brain parcellation performed using GIF, not included in this repository).
(Brain parcellation performed using GIF, not included in this repository).
This package uses NiftyNet, which is built on top of TensorFlow, so first of all you need to follow the instructions to setup your NVIDIA GPU.
While it is possible to perform a segmentation without a GPU, inference of one large volume (512 × 512 × 394 voxels) using the default parameters takes one hour using CPU only and around 90 seconds using a GPU.
Using conda is recommended:
$ conda create -n "vesseg" python=3.6 -y # tensorflow doesn't support python 3.7 yet
$ conda activate "vesseg"
(vesseg) $ git clone "https://github.com/fepegar/vesseg.git" --depth 1
(vesseg) $ pip install --editable "./vesseg"$ conda activate vesseg
(vesseg) $ vesseg dsa_image.nii.gz dsa_vessels_seg.nii.gz
(vesseg) $ bin2mesh dsa_vessels_seg.nii.gz dsa_vessels_mesh.vtpTested on Linux and macOS.