Volume Segmentation Tool with GUI
This repository holds the script for Volume Segmentation Tool and its Gradio-based Webui.
Support Operating Systems: Windows and Linux.
MacOS support is a work in process.
About the Tool
Volume Segmentation Tool is a python based tool that utilizes deep learning to perform volumetric electron microscopy image segmentations.
Based on Pytorch backend.
The desired images are 8bit or 16bit grey scale 3D tif as commonly generated by electron microscopes.
This tool does not support 2D image segmentation, nor colorful image segmentation. In which case you should consider ilastik or Trainable Weka Segmentation.
Advantages
- High efficiency
- Custom implemented augmentation specialised in volumetric images
- Process many depth slices all once, rather than slice by slice
- Support fp16 training
- Versatile
- Semantic Segmentation & Instance Segmentation
- Isotropic & anisotropic images
- Customisable network type, size and depth
- Support AMD GPU (only under Linux)
- Easy to use
- One click installation script
- Graphical User Interface
- Visualisation tools for data augmentation and network activations
- Visualise network output on the fly
- TensorBoard Logging & Can export as Excel spreadsheet
Limitations
- As for now, MacOS support is a work in process
- Does not support 2D images, nor images with colours (grey scale only)
- Since it's based on Deep Learning, the tool needs to be used with a discrete GPU
- It's possible to run this tool without a discrete GPU, however it will run extremely slowly
- Recommended minimal GPU requirement: 4GB of Video Memory, made by Nvidia or AMD
- Since it's based on Deep Learning, the user has to create training samples to train the network before it could perform segmentation
- For more information, please see Tutorials.
Installation
-
Install Python 3.10.
- During the installation process, ensure that you select the option to add Python to the 'PATH' environment variable.
-
Install Git.
- If you are using Linux, it's highly likely you can skip this step.
-
Open a terminal and navigate to the desired installation directory.
-
Clone the repository by running the following command:
git clone https://github.com/fgdfgfthgr-fox/Volume_Seg_Tool.git
-
Run the corresponding "install_dependencies" script for your system.
- For Windows, it would be install_dependencies_Windows.bat
-
Wait for the script to finish, which could take a while depends on your network speed.
Starting GUI
- Run the corresponding "start_WebUI" script for your system. This would open up a terminal.
- For Windows, it would be start_WebUI_Windows.bat
- Your web browser should automatically open up a "website" with url "127.0.0.1:7860" or something similar.
Tutorials
Please see the Wiki (Mostly WIP).
Credits
This tool was developed under the scholarship funding from AgResearch, and was helped by the members of the Bostina Lab of the University of Otago. As well as Lech Szymanski from the Computer Science department.
The example dataset included in this repository was collected by Vincent Casser.
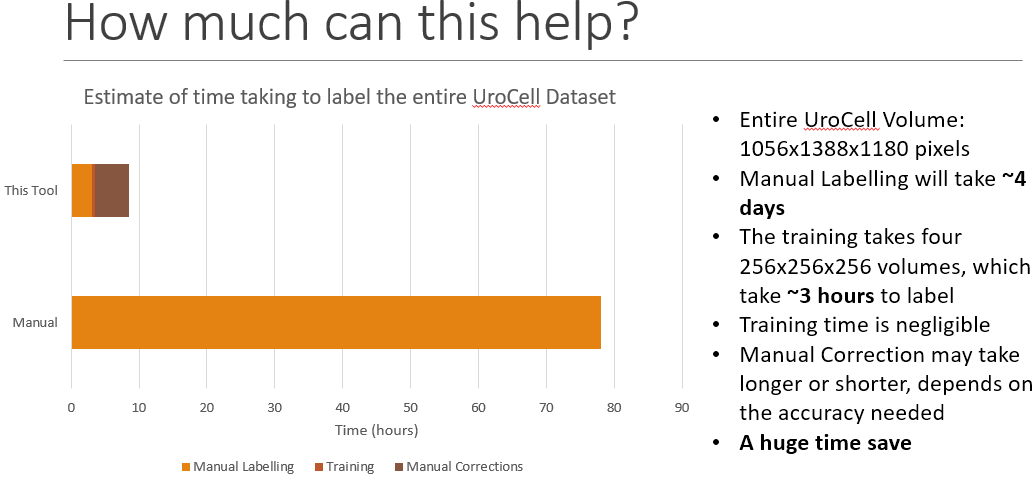
The UroCell Dataset was heavily used during the development of the tool.
Special thanks to YunBo Wang from Xidian University, who gave me exceptional helps at the early stage of development.