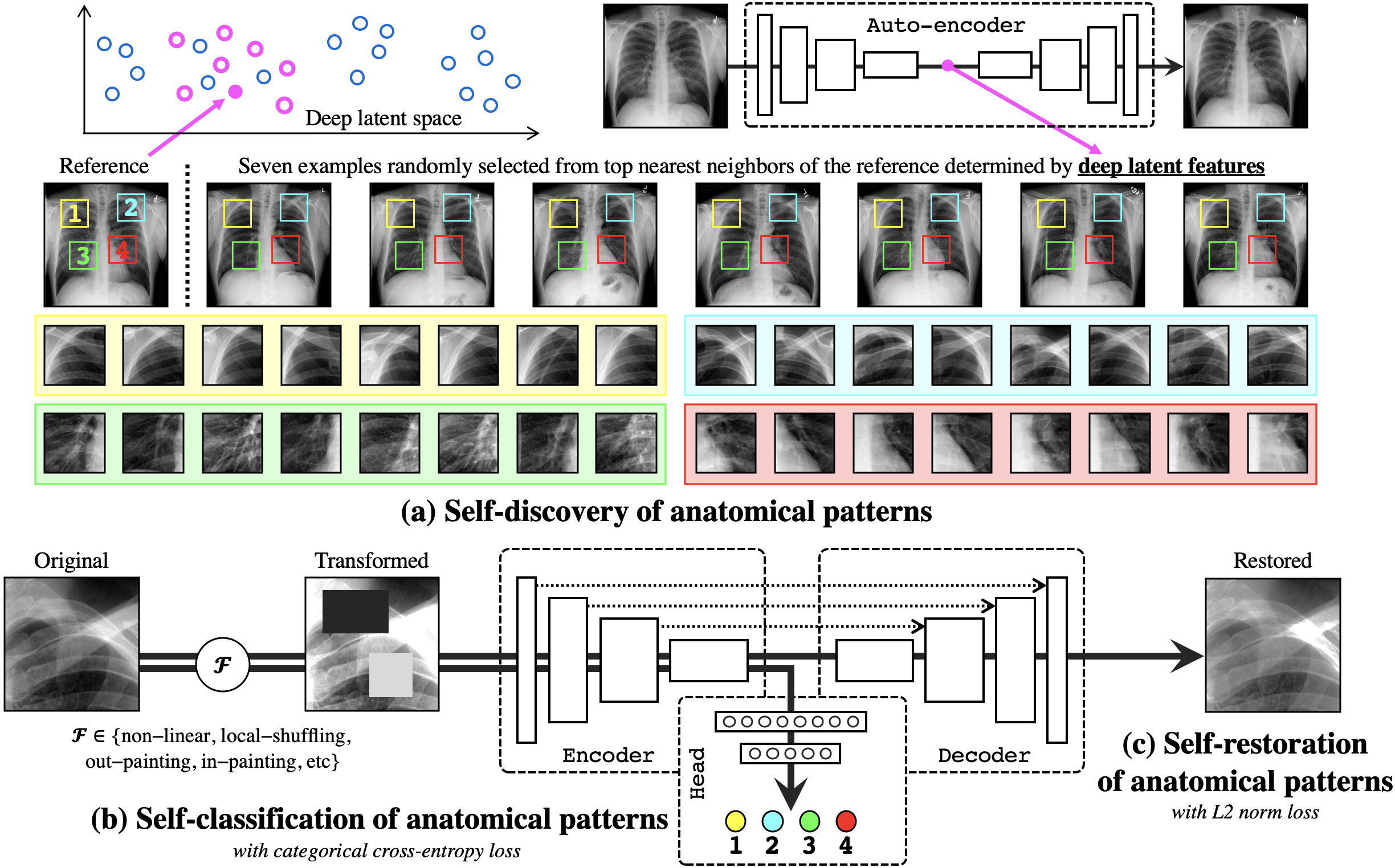
We have trained deep models to learn semantically enriched visual representation by self-discovery, self-classification, and self-restoration of the anatomy underneath medical images, resulting in a semantics-enriched, general-purpose, pre-trained 3D model, named Semantic Genesis . Not only does our self-supervised framework outperform existing methods, but also it can be used as an add-on to improve existing representation learning methods. This key contribution enables many representation learning methods to learn semantics-enriched representations from unlabeled medical images, a remarkable achievement in general-purpose representation learning.
This repository provides the official implementation of training Semantic Genesis as well as using the pre-trained Semantic Genesis in the following paper:
Learning Semantics-enriched Representation via Self-discovery, Self-classification, and Self-restoration
Fatemeh Haghighi1, Mohammad Reza Hosseinzadeh Taher1, Zongwei Zhou1, Michael B. Gotway2, Jianming Liang1
1Arizona State University, 2 Mayo Clinic
International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2020
Paper | Code | Poster | Slides | Graphical abstract | Talk (YouTube, YouKu)
Transferable Visual Words: Exploiting the Semantics of Anatomical Patterns for Self-supervised Learning
Fatemeh Haghighi1, Mohammad Reza Hosseinzadeh Taher1,Zongwei Zhou1,Michael B. Gotway2, Jianming Liang1
1 Arizona State University, 2Mayo Clinic
IEEE Transactions on Medical Imaging (TMI)
paper | code
- Learning semantics through our proposed self-supervised learning framework enriches existing self-supervised methods
- Semantic Genesis outperforms:
- 3D models trained from scratch
- 3D self-supervised pre-trained models
- 3D supervised pre-trained models
Credit to superbar by Scott Lowe for Matlab code of superbar.
If you use this code or use our pre-trained weights for your research, please cite our paper:
@InProceedings{haghighi2020learning,
author="Haghighi, Fatemeh and Hosseinzadeh Taher, Mohammad Reza and Zhou, Zongwei and Gotway, Michael B. and Liang, Jianming",
title="Learning Semantics-Enriched Representation via Self-discovery, Self-classification, and Self-restoration",
booktitle="Medical Image Computing and Computer Assisted Intervention -- MICCAI 2020",
year="2020",
publisher="Springer International Publishing",
address="Cham",
pages="137--147",
isbn="978-3-030-59710-8",
url="https://link.springer.com/chapter/10.1007%2F978-3-030-59710-8_14"
}
@ARTICLE{haghighi2021transferable,
author={Haghighi, Fatemeh and Taher, Mohammad Reza Hosseinzadeh and Zhou, Zongwei and Gotway, Michael B. and Liang, Jianming},
journal={IEEE Transactions on Medical Imaging},
title={Transferable Visual Words: Exploiting the Semantics of Anatomical Patterns for Self-Supervised Learning},
year={2021},
volume={40},
number={10},
pages={2857-2868},
doi={10.1109/TMI.2021.3060634}}
This research has been supported partially by ASU and Mayo Clinic through a Seed Grant and an Innovation Grant, and partially by the NIH under Award Number R01HL128785. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work has utilized the GPUs provided partially by the ASU Research Computing and partially by the Extreme Science and Engineering Discovery Environment (XSEDE) funded by the National Science Foundation (NSF) under grant number ACI-1548562. We thank Zuwei Guo for implementing Rubik's cube, M. M. Rahman Siddiquee for examining NiftyNet, and Jiaxuan Pang for evaluating I3D. The content of this paper is covered by patents pending.
Released under the ASU GitHub Project License.