TL.DR version:
I was able to predict whether the loan is default(including charged off) like most kernals regarding this dataset with 99.96% accuracy.
But LendingClub also has a internal 'Grade' like A, B, C, D and 'Subgrade' like A1, B2, C3 for its customers, which is more ineresting as a metric of business decision than loan status We also reverse enginner LendingClub's 'Grade' and 'Subgrade' rating system with 99.6% accuracy, 13minutes of traiinng with boosting and 99.86% accuracy, 25 minute of trainig with boosting respectively.
I also used Fast.AI's tabular module for training, which achieved 97% and 87% accuracy with GPU acceleration. Data cleaning was proven to be critical: before data cleaning and feature enginnering, the accuracy is less than 20%.
To make LightGBM training faster, I attemped to setup LightBGM but met many problems. A pull request is submitted with potentially more to come to Microsoft's LightBGM team.
Thanks for reading! I would really appreciate any feedback.
Happy coding, Alex Li alex@alexli.me
# The Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("./input"))
# Any results you write to the current directory are saved as output.
https://www.kaggle.com/wendykan/lending-club-loan-data
##Reading the dataset
df = pd.read_csv("./input/loan_untouched.csv", low_memory=False)
df.head(3)Note that it has consumed about 10Gb of memory, 11.8GB specifically. This is a sign that we need to be very careful about memory mangagement, especially in a low-resource setting.
Let us check some basics about the dataset. First about its size and then about its dimensions.
df.shape df.columnsWhat does the various columns mean? We can rely on the dictionary provided by the Lending Street. We use Pandas to read the xlsx file, along with display style formatting
df_description = pd.read_excel('./input/LCDataDictionary.xlsx').dropna()
df_description.style.set_properties(subset=['Description'], **{'width': '1000px'})We start out with the metric of the game: loan status
df["loan_status"].value_counts()df["loan_status"].value_counts().plot(kind='bar');We see that there are only 31 default cases. In the light that harges off means that the consumer has defaulted for long enough time such that the creditors has 'given up' in getting the money back, we include them in our default cases.
target_list = [1 if i=='Default' or i=='Charged Off' else 0 for i in df['loan_status']]
df['TARGET'] = target_list
df['TARGET'].value_counts()Even after including the charges off cases, we have a class imbalance issue: the 'positive' cases of defaults are much less frequent than the 'negative' non-default cases. Later in training, we need to be careful about this.
# loan_status cross
loan_status_cross = pd.crosstab(df['addr_state'], df['loan_status']).apply(lambda x: x/x.sum() * 100)
number_of_loanstatus = pd.crosstab(df['addr_state'], df['loan_status'])
# Round our values
loan_status_cross['Charged Off'] = loan_status_cross['Charged Off'].apply(lambda x: round(x, 2))
loan_status_cross['Default'] = loan_status_cross['Default'].apply(lambda x: round(x, 2))
loan_status_cross['Does not meet the credit policy. Status:Charged Off'] = loan_status_cross['Does not meet the credit policy. Status:Charged Off'].apply(lambda x: round(x, 2))
loan_status_cross['In Grace Period'] = loan_status_cross['In Grace Period'].apply(lambda x: round(x, 2))
loan_status_cross['Late (16-30 days)'] = loan_status_cross['Late (16-30 days)'].apply(lambda x: round(x, 2))
loan_status_cross['Late (31-120 days)'] = loan_status_cross['Late (31-120 days)'].apply(lambda x: round(x, 2))
number_of_loanstatus['Total'] = number_of_loanstatus.sum(axis=1)
# number_of_badloans
number_of_loanstatusActually, after we set the target column, we can drop the loan_status column, as a role of thumb
df.drop('loan_status',axis=1,inplace=True)df.info()from plotly import tools
import chart_studio.plotly as py
import plotly.figure_factory as ff
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
init_notebook_mode(connected=True)plt.figure(figsize=(8, 5))
sns.barplot(y=df.term.value_counts(), x=df.term.value_counts().index)
plt.xticks(rotation=0)
plt.title("Loan's Term Distribution")
plt.ylabel("Count")plt.figure(figsize=(18, 5))
sns.barplot(y=df.grade.value_counts(), x=df.grade.value_counts().index)
plt.xticks(rotation=0)
plt.title("Grade Distribution")
plt.ylabel("Count")plt.figure(figsize=(18, 10))
sns.barplot(y=df.sub_grade.value_counts(), x=df.sub_grade.value_counts().index)
plt.xticks(rotation=0)
plt.title("Subgrade Distribution")
plt.ylabel("Count")plt.figure(figsize=(18, 10))
sns.barplot(y=df.purpose.value_counts(), x=df.purpose.value_counts().index)
plt.xticks(rotation=0)
plt.title("Purpose Distribution")
plt.ylabel("Count")
plt.xticks(rotation=30)plt.figure(figsize=(18, 10))
sns.barplot(y=df.emp_title.value_counts()[:15], x=df.emp_title.value_counts()[:15].index)
plt.xticks(rotation=0)
plt.title("Top 15 most popular employment title")
plt.ylabel("Count")
plt.xticks(rotation=30)plt.figure(figsize = (14,6))
sns.barplot(x='grade', y='loan_amnt',
data=df)
g.set_xticklabels(g.get_xticklabels(),rotation=90)
g.set_xlabel("Duration Distribuition", fontsize=15)
g.set_ylabel("Mean amount", fontsize=15)
g.set_title("Loan Amount by Grades", fontsize=20)
plt.legend(loc=1)
plt.show()def null_values(df):
mis_val = df.isnull().sum()
mis_val_percent = 100 * df.isnull().sum() / len(df)
mis_val_table = pd.concat([mis_val, mis_val_percent], axis=1)
mis_val_table_ren_columns = mis_val_table.rename(
columns = {0 : 'Missing Values', 1 : '% of Total Values'})
mis_val_table_ren_columns = mis_val_table_ren_columns[
mis_val_table_ren_columns.iloc[:,1] != 0].sort_values(
'% of Total Values', ascending=False).round(1)
print ("Dataframe has " + str(df.shape[1]) + " columns.\n"
"There are " + str(mis_val_table_ren_columns.shape[0]) +
" columns that have missing values.")
return mis_val_table_ren_columnsmissing_values = null_values(df)missing_valuesMore specifically, we can look at the missing values for float64 type of columns, sorted by the percentage of missing
df.select_dtypes(include=['float64']).describe()\
.T.assign(missing = df.apply(lambda x : (1 - x.count() /len(x)))).sort_values(by = 'missing',ascending = False)We can also look at the missing values for object type of columns, sorted by the percentage of missing
df.select_dtypes(include=['O']).describe()\
.T.assign(missing = df.apply(lambda x : (1 - x.count() /len(x)))).sort_values(by = 'missing',ascending = False)We decides to drop the columns where 25% or more values is missing
df.dropna(
axis=1, thresh=int(0.75 * len(df)),
inplace=True)We can take a look at the new dataframe
df.describe().T.assign(
missing_pct=df.apply(
lambda x: (1 - x.count() / len(x)))).sort_values(
by='missing_pct', ascending=False)Here we look at each columns in the above, we can just take a look at the top columns and analyze the financial context of each column.
The basic logic below is that we assume a 'good' population that follow a summary statistics that makes sense simply by common sense, like positive annual income (instead of 0 income)
mths_since_recent_inq: should be filled with mean
num_tl_120dpd_2m: Number of accounts currently 120 days past due: should be filled with mean
mo_sin_old_il_acct:Months since oldest bank installment account opened. Should be filled with mean
bc_util: Ratio of total current balance to high credit/credit limit for all bankcard accounts: Should be filled with mean
percent_bc_gt_75: Percentage of all bankcard accounts > 75% of limit: should be filled with zero
bc_open_to_buy: Total open to buy on revolving bankcards. Should be filled with zero
mths_since_recent_bc: Months since most recent bankcard account opened: Should be filled with zero
pct_tl_nvr_dlq: Percent of trades never delinquent: should be filled with zero
avg_cur_bal: Average current balance of all accounts: should be filled with zero
mo_sin_rcnt_rev_tl_op: Months since most recent revolving account opened: should be filled with zero
mo_sin_old_rev_tl_op: Months since most recent revolving account opened: should be filled with mean
num_rev_accts:Number of revolving accounts: should be filled with zero
tot_cur_bal: Total current balance of all accounts: should be filled with zero
num_il_tl: Number of installment accounts: should be filled with zero
tot_coll_amt: Total collection amounts ever owed: Should be filled with zero
num_accts_ever_120_pd:Number of accounts ever 120 or more days past due: Should be filled with zero
cols_fill_with_mean = ['mths_since_recent_inq',
'num_tl_120dpd_2m',
'mo_sin_old_il_acct',
'bc_util',
'mo_sin_old_rev_tl_op',
'revol_util', 'open_acc', 'total_acc',
'annual_inc']for col in cols_fill_with_mean:
mean_of_col = df[col].mean()
df[col] = df[col].fillna(mean_of_col)df['emp_title']df['emp_title'].value_counts()df['title']df['title'].value_counts()df['zip_code']For emp_title and title, we notice that there are too many entries that appears only once. If we were to do one-hot encoding, they would add unnecessary complexity to our parameter space. We do recognize that if we have more time, we should convert low-frequency title and emp-titles into one category while keeping high-frequency titles. For zip_code, since the last two digits are missing, it also will add necessary complexity in parameter space while not contributing to analysis.
df.drop(['emp_title','title','zip_code'],axis=1,inplace=True)df.shapeAs the last step we take a look at the columns with missing values after basic cleaning and dropping. We need to specifiy object columns and number columns
df.select_dtypes(include=['O']).describe()\
.T.assign(missing = df.apply(lambda x : (1 - x.count() /len(x)))).sort_values(by = 'missing',ascending = False)Note that only the first four columns have value missing.
Since there are only a few columns
df['emp_length']Let us fill in the null values with 0 assuming the applicant does not work and consequently does not report their employment length. And then we convert the format of the column from object to
df['emp_length'].fillna(value=0,inplace=True)
df['emp_length'].replace(to_replace='[^0-9]+', value='', inplace=True, regex=True)
df['emp_length'].value_counts().sort_values().plot(kind='barh',figsize=(18,8))
plt.title('Number of loans distributed by Employment Years',fontsize=20)
plt.xlabel('Number of loans',fontsize=15)
plt.ylabel('Years worked',fontsize=15);Also, there are four month/date object that we can simply use pandas built-in toolbox to convert to datetime object. Instead of filling the null values with zero values, we can fill with the most frequenct value.
df['last_pymnt_d'].value_counts().sort_values().tail()We know we want to fill the missing value with Feb-2019 as the most frequent date for last_pymnt_d
df['last_pymnt_d']= pd.to_datetime(df['last_pymnt_d'].fillna('2016-09-01'))df['last_credit_pull_d'].value_counts().sort_values().tail()We know we want to fill the missing value with Feb-2019 as the most frequent date for last_credit_pull_d
df['last_credit_pull_d']= pd.to_datetime(df['last_credit_pull_d'].fillna("2016-09-01"))df['earliest_cr_line'].value_counts().sort_values().tail()We know we want to fill the missing value with Sep-2004 as the most frequent date for earliest_cr_line
df['earliest_cr_line']= pd.to_datetime(df['earliest_cr_line'].fillna('2004-09-01'))Now we shall use label encoding to encode the object columns with two unique values or less
We know we want to fill the missing value with Mar 2016 as the most frequent date for issue_d
df['issue_d'].value_counts().sort_values().tail()df['issue_d']= pd.to_datetime(df['issue_d'].fillna("2016-03-01"))
Save the dataframe
df.to_pickle('Before_datetime')import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as snsdf = pd.read_pickle('Before_datetime')df.select_dtypes(exclude=[np.number,object]).head()Two metric we can feature enginnering is:
- The months between issue date and last payment date, which is how long the loan has been around
- The months between earliest credit line and the last credit pull date, which is similar to the length of credit history
df['months_of_loan']= (df['last_pymnt_d']-df['issue_d'])/np.timedelta64(1,'M')df['months_of_loan']=df['months_of_loan'].astype(int)df['months_of_credit_hisotory']= (df['last_credit_pull_d']-df['earliest_cr_line'])/np.timedelta64(1,'M')df['months_of_credit_hisotory']=df['months_of_credit_hisotory'].astype(int)df.info()df.select_dtypes(object).head()We see that the term of the loan is still in string/object format, let us convert that into int
df['term'].replace(to_replace='[^0-9]+', value='', inplace=True, regex=True)
df['term']=df['term'].astype(int)Check if 'term' is properly converted
df['term'].head()We remember that we converted emp_length. Check if it is in int format, if not, convert it to int
df['emp_length'].head()df['emp_length']= df['emp_length'].astype(int)df.select_dtypes(object).columnsdf.to_pickle('ReadyforModelling')**Note that ideally, we would like to model three metrics **
- Loan Status (We converted it into 'target' column), the lending club's final decision. However We do recognize that Lending Club's business case is unique in terms of P2P lending. We should also model other parameters.
- 'Grade'. This is Lending Club's secret sauce of assigning a grade to a borrower. More universal and thus more interesting to model.
- 'Subgrade'. 'Grade' metric is added by another single digit number to further distingusih between different customers
When we model loan status, it is considered common to include 'Grade' and 'Subgrade'. But when we model 'Grade' and 'Subgrade', it is debetable to include Loan Status or not However, we know for certain that 'Subgrade' should not be a metric for 'Grade' modelling And reversely, 'Grade' should not be a metric for 'Subgrade'
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score, classification_report, confusion_matrixdf=pd.read_pickle('ReadyforModelling')Label encoding the binary objects
from sklearn import preprocessing
count = 0
for col in df:
if df[col].dtype == 'object':
if len(list(df[col].unique())) <= 2:
le = preprocessing.LabelEncoder()
df[col] = le.fit_transform(df[col])
count += 1
print (col)
print('%d columns were label encoded.' % count)Now we can use get_dummies to one hot encode all the rest categorical columns. We just need to convert the datetime objects back to strings
df['last_pymnt_d'] = df['last_pymnt_d'].dt.strftime('%Y-%m')
df['issue_d'] =df['issue_d'].dt.strftime('%Y-%m')
df['last_credit_pull_d']=df['last_credit_pull_d'].dt.strftime('%Y-%m')
df['earliest_cr_line']=df['earliest_cr_line'].dt.strftime('%Y-%m')df = pd.get_dummies(df)And we can drop the remaining null values now
df.dropna(inplace=True)import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score, classification_report, confusion_matrix
import os
import gcdef print_score(clf, X_train, y_train, X_test, y_test, train=True):
if train:
print("Train Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_train, clf.predict(X_train))))
print("Classification Report: \n {}\n".format(classification_report(y_train, clf.predict(X_train))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_train, clf.predict(X_train))))
res = cross_val_score(clf, X_train, y_train, cv=10, scoring='accuracy')
print("Average Accuracy: \t {0:.4f}".format(np.mean(res)))
print("Accuracy SD: \t\t {0:.4f}".format(np.std(res)))
elif train==False:
print("Test Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_test, clf.predict(X_test))))
print("Classification Report: \n {}\n".format(classification_report(y_test, clf.predict(X_test))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_test, clf.predict(X_test))))
from sklearn.model_selection import train_test_splitX_train, X_test, y_train, y_test = train_test_split(df.drop('TARGET',axis=1),df['TARGET'],test_size=0.15,random_state=101)from sklearn.preprocessing import StandardScalersc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test=sc.transform(X_test)/opt/anaconda3/lib/python3.7/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64, float64 were all converted to float64 by StandardScaler.
return self.partial_fit(X, y)
/opt/anaconda3/lib/python3.7/site-packages/sklearn/base.py:462: DataConversionWarning: Data with input dtype int64, float64 were all converted to float64 by StandardScaler.
return self.fit(X, **fit_params).transform(X)
/opt/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:3: DataConversionWarning: Data with input dtype int64, float64 were all converted to float64 by StandardScaler.
This is separate from the ipykernel package so we can avoid doing imports until
We can create a baseline for accuracty and recall using logistics regression
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression(C = 0.0001,random_state=21)
log_reg.fit(X_train, y_train)/opt/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/logistic.py:433: FutureWarning: Default solver will be changed to 'lbfgs' in 0.22. Specify a solver to silence this warning.
FutureWarning)
LogisticRegression(C=0.0001, class_weight=None, dual=False,
fit_intercept=True, intercept_scaling=1, max_iter=100,
multi_class='warn', n_jobs=None, penalty='l2', random_state=21,
solver='warn', tol=0.0001, verbose=0, warm_start=False)
print_score(log_reg, X_train, y_train, X_test, y_test, train=False)Test Result:
accuracy score: 0.9786
Classification Report:
precision recall f1-score support
0 0.98 1.00 0.99 287182
1 1.00 0.82 0.90 37342
micro avg 0.98 0.98 0.98 324524
macro avg 0.99 0.91 0.94 324524
weighted avg 0.98 0.98 0.98 324524
Confusion Matrix:
[[287048 134]
[ 6825 30517]]
**Note that for our business case, we would like to reduce the top right number in the confusion matrix which is false postive: we think that the applicant is a good loan borrower but he/she is not. Compared to the bottom left number, which is false negative, we as a business does not loss when we reject a good applicant, at least not as much as giving loan to a bad loaner. **
from sklearn.ensemble import RandomForestClassifierclf_rf = RandomForestClassifier(n_estimators=40, random_state=21)
clf_rf.fit(X_train, y_train)RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=40, n_jobs=None,
oob_score=False, random_state=21, verbose=0, warm_start=False)
print_score(clf_rf, X_train, y_train, X_test, y_test, train=False)Test Result:
accuracy score: 0.9968
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 287182
1 1.00 0.97 0.99 37342
micro avg 1.00 1.00 1.00 324524
macro avg 1.00 0.99 0.99 324524
weighted avg 1.00 1.00 1.00 324524
Confusion Matrix:
[[287182 0]
[ 1052 36290]]
- Train weak classifiers
- Add them to a final strong classifier by weighting. Weighting by accuracy (typically)
- Once added, the data are reweighted
- Misclassified samples gain weight
- Algo is forced to learn more from misclassified samples
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import roc_auc_score, roc_curve
from sklearn.model_selection import KFold, StratifiedKFold
from lightgbm import LGBMClassifier
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import gc
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score, classification_report, confusion_matrixdf=pd.read_pickle('ReadyforModelling')def kfold_lightgbm(train_df, num_folds, stratified = False):
print("Starting LightGBM. Train shape: {}".format(train_df.shape))
# Cross validation model
if stratified:
folds = StratifiedKFold(n_splits= num_folds, shuffle=True, random_state=47)
else:
folds = KFold(n_splits= num_folds, shuffle=True, random_state=47)
oof_preds = np.zeros(train_df.shape[0])
feature_importance_df = pd.DataFrame()
feats = [f for f in train_df.columns if f not in ['TARGET']]
# Splitting the training set into folds for Cross Validation
for n_fold, (train_idx, valid_idx) in enumerate(folds.split(train_df[feats], train_df['TARGET'])):
train_x, train_y = train_df[feats].iloc[train_idx], train_df['TARGET'].iloc[train_idx]
valid_x, valid_y = train_df[feats].iloc[valid_idx], train_df['TARGET'].iloc[valid_idx]
# LightGBM parameters found by Bayesian optimization
clf = LGBMClassifier(
nthread=12,#previous number 4
n_estimators=1000, # Previous number 10000
learning_rate=0.02,
num_leaves=32,
colsample_bytree=0.9497036,
subsample=0.8715623,
max_depth=8,
reg_alpha=0.04,
reg_lambda=0.073,
min_split_gain=0.0222415,
min_child_weight=40,
silent=-1,
verbose=-1
#device= 'gpu',gpu_platform_id= 0,gpu_device_id= 0
)
# Fitting the model and evaluating by AUC
clf.fit(train_x, train_y, eval_set=[(train_x, train_y), (valid_x, valid_y)],
eval_metric= 'auc', verbose= 1000, early_stopping_rounds= 200)
print_score(clf, train_x, train_y, valid_x, valid_y, train=False)
# Dataframe holding the different features and their importance
fold_importance_df = pd.DataFrame()
fold_importance_df["feature"] = feats
fold_importance_df["importance"] = clf.feature_importances_
fold_importance_df["fold"] = n_fold + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
# Freeing up memory
del clf, train_x, train_y, valid_x, valid_y
gc.collect()
display_importances(feature_importance_df)
return feature_importance_dfdef print_score(clf, X_train, y_train, X_test, y_test, train=True):
if train:
print("Train Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_train, clf.predict(X_train))))
print("Classification Report: \n {}\n".format(classification_report(y_train, clf.predict(X_train))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_train, clf.predict(X_train))))
res = cross_val_score(clf, X_train, y_train, cv=10, scoring='accuracy')
print("Average Accuracy: \t {0:.4f}".format(np.mean(res)))
print("Accuracy SD: \t\t {0:.4f}".format(np.std(res)))
elif train==False:
print("Test Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_test, clf.predict(X_test))))
print("Classification Report: \n {}\n".format(classification_report(y_test, clf.predict(X_test))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_test, clf.predict(X_test)))) def display_importances(feature_importance_df_):
cols = feature_importance_df_[["feature", "importance"]].groupby("feature").mean().sort_values(by="importance", ascending=False)[:40].index
best_features = feature_importance_df_.loc[feature_importance_df_.feature.isin(cols)]
plt.figure(figsize=(15, 12))
sns.barplot(x="importance", y="feature", data=best_features.sort_values(by="importance", ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
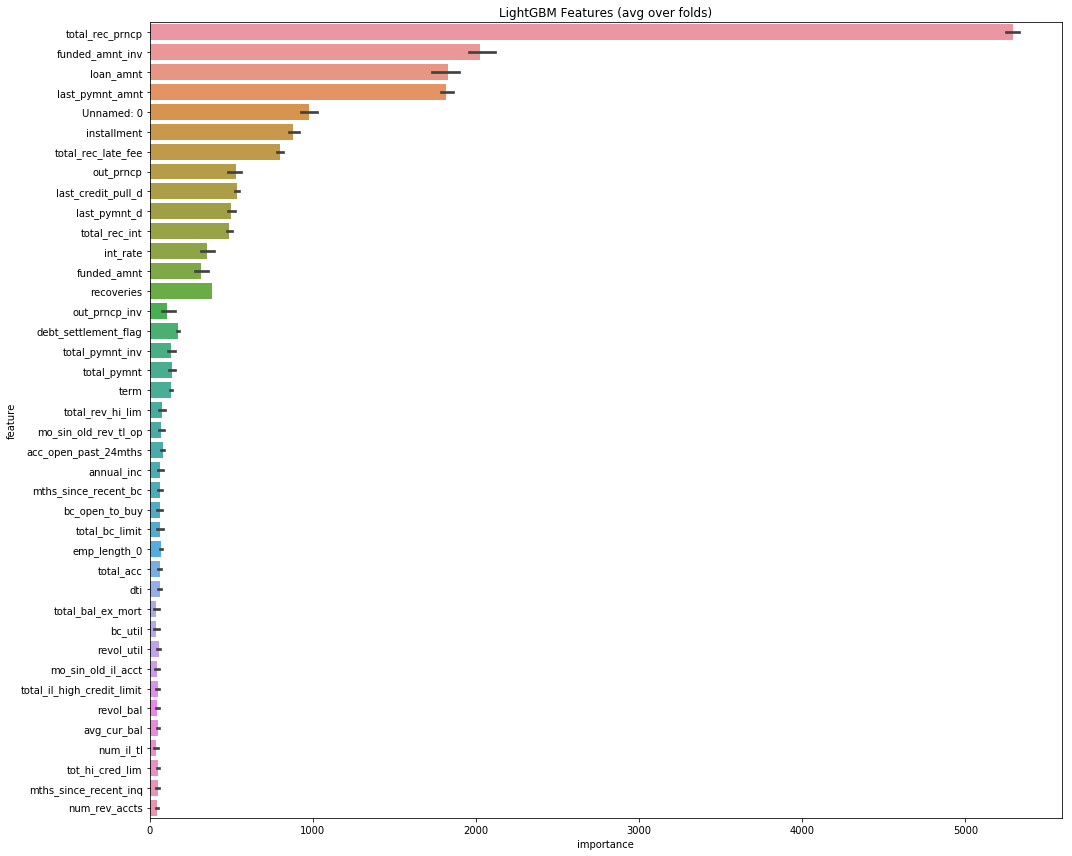
plt.savefig('lgbm_importances.png')feat_importance = kfold_lightgbm(df, num_folds= 5, stratified= False)Starting LightGBM. Train shape: (2163488, 343)
Training until validation scores don't improve for 200 rounds.
[1000] training's binary_logloss: 0.00154127 training's auc: 0.999953 valid_1's binary_logloss: 0.00183299 valid_1's auc: 0.999887
Did not meet early stopping. Best iteration is:
[1000] training's binary_logloss: 0.00154127 training's auc: 0.999953 valid_1's binary_logloss: 0.00183299 valid_1's auc: 0.999887
Test Result:
accuracy score: 0.9996
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 383090
1 1.00 1.00 1.00 49608
micro avg 1.00 1.00 1.00 432698
macro avg 1.00 1.00 1.00 432698
weighted avg 1.00 1.00 1.00 432698
Confusion Matrix:
[[383090 0]
[ 168 49440]]
Training until validation scores don't improve for 200 rounds.
[1000] training's binary_logloss: 0.00153117 training's auc: 0.999951 valid_1's binary_logloss: 0.00180118 valid_1's auc: 0.999917
Did not meet early stopping. Best iteration is:
[1000] training's binary_logloss: 0.00153117 training's auc: 0.999951 valid_1's binary_logloss: 0.00180118 valid_1's auc: 0.999917
Test Result:
accuracy score: 0.9996
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 382983
1 1.00 1.00 1.00 49715
micro avg 1.00 1.00 1.00 432698
macro avg 1.00 1.00 1.00 432698
weighted avg 1.00 1.00 1.00 432698
Confusion Matrix:
[[382983 0]
[ 167 49548]]
Training until validation scores don't improve for 200 rounds.
[1000] training's binary_logloss: 0.00154649 training's auc: 0.999949 valid_1's binary_logloss: 0.00173747 valid_1's auc: 0.999929
Did not meet early stopping. Best iteration is:
[1000] training's binary_logloss: 0.00154649 training's auc: 0.999949 valid_1's binary_logloss: 0.00173747 valid_1's auc: 0.999929
Test Result:
accuracy score: 0.9996
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 382891
1 1.00 1.00 1.00 49807
micro avg 1.00 1.00 1.00 432698
macro avg 1.00 1.00 1.00 432698
weighted avg 1.00 1.00 1.00 432698
Confusion Matrix:
[[382890 1]
[ 177 49630]]
Training until validation scores don't improve for 200 rounds.
[1000] training's binary_logloss: 0.0015465 training's auc: 0.999951 valid_1's binary_logloss: 0.00164619 valid_1's auc: 0.999948
Did not meet early stopping. Best iteration is:
[1000] training's binary_logloss: 0.0015465 training's auc: 0.999951 valid_1's binary_logloss: 0.00164619 valid_1's auc: 0.999948
Test Result:
accuracy score: 0.9996
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 382931
1 1.00 1.00 1.00 49766
micro avg 1.00 1.00 1.00 432697
macro avg 1.00 1.00 1.00 432697
weighted avg 1.00 1.00 1.00 432697
Confusion Matrix:
[[382931 0]
[ 169 49597]]
Training until validation scores don't improve for 200 rounds.
[1000] training's binary_logloss: 0.00151123 training's auc: 0.999956 valid_1's binary_logloss: 0.00182654 valid_1's auc: 0.999912
Did not meet early stopping. Best iteration is:
[1000] training's binary_logloss: 0.00151123 training's auc: 0.999956 valid_1's binary_logloss: 0.00182654 valid_1's auc: 0.999912
Test Result:
accuracy score: 0.9996
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 383173
1 1.00 1.00 1.00 49524
micro avg 1.00 1.00 1.00 432697
macro avg 1.00 1.00 1.00 432697
weighted avg 1.00 1.00 1.00 432697
Confusion Matrix:
[[383173 0]
[ 169 49355]]
/opt/anaconda3/lib/python3.7/site-packages/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval
We are relative excited by the 0 false positve value in the confusion matrix. This means that we did not borrow money to anyone who was going to default
Admittedly, due to time constraints (time conflicts with other job commitments) I was unable to produce complete analysis of Grade and Subgrade. The following is the fastai/pytorch code. It shall be run in a new envionment with Fastai installed with GPU/CUDA support
from fastai.tabular import * # Fast AI tabular packagepd.read_csv("./input/loan_untouched.csv", low_memory=False)procs = [FillMissing, Normalize]To split our data into training and validation sets, we use valid indexes
valid_idx = range(len(df)-2000, len(df))Let us use the helper function that returns column names of cont and cat variables from given df.
Now we're ready to pass this information to TabularDataBunch.from_df to create the DataBunch that we'll use for training. Once we have our data ready in a DataBunch, we just need to create a model to then define a Learner and start training. The fastai library has a flexible and powerful TabularModel in models.tabular. To use that function, we just need to specify the embedding sizes for each of our categorical variables.
dep_var='grade'
cont_list, cat_list = cont_cat_split(df=df, max_card=2000, dep_var='TARGET')
cont_list, cat_listdata = TabularDataBunch.from_df(path, df, dep_var, valid_idx=valid_idx, procs=procs, cat_names=cat_list)
print(data.train_ds.cont_names) # `cont_names` defaults to: set(df)-set(cat_names)-{dep_var}learn = tabular_learner(data, layers=[200,100], metrics=accuracy)learn.model_dir=Path("../")lr_find(learn)learn.fit_one_cycle(1, 1e-6)import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as snsdf = pd.read_pickle('ReadyforModelling')df['last_pymnt_d'] = df['last_pymnt_d'].dt.strftime('%Y-%m')
df['issue_d'] =df['issue_d'].dt.strftime('%Y-%m')
df['last_credit_pull_d']=df['last_credit_pull_d'].dt.strftime('%Y-%m')
df['earliest_cr_line']=df['earliest_cr_line'].dt.strftime('%Y-%m')df.info()** Let us convert the grades and subgrades into numerical categories **
factor_grade = pd.factorize(df['grade'])
factor_gradedf['grade'] = factor_grade[0]factor_subgrade = pd.factorize(df['sub_grade'])
factor_subgradedf['sub_grade'] = factor_subgrade[0]df.select_dtypes(exclude=[np.number]).head()We see that the term of the loan is still in string/object format, let us convert that into int
df['term'].replace(to_replace='[^0-9]+', value='', inplace=True, regex=True)
df['term']=df['term'].astype(int)Check if 'term' is properly converted
df['term'].head()We remember that we converted emp_length. Check if it is in int format, if not, convert it to int
df['emp_length'].head()df['emp_length']= df['emp_length'].astype(int)df.select_dtypes(object).columnsfrom sklearn import preprocessing
count = 0
for col in df:
if df[col].dtype == 'object':
if len(list(df[col].unique())) <= 2:
le = preprocessing.LabelEncoder()
df[col] = le.fit_transform(df[col])
count += 1
print (col)
print('%d columns were label encoded.' % count)df.select_dtypes(object).columnsFor object columsn with more than 2 classes, we shall use one hot encoding instead. If we were to apply label encoding for multi class objects, we would potentailly make the system think that there is order between 0, 1, and 2, etc. as representation type.
df = pd.get_dummies(df)
print(df.shape)We now fill in the null values with zero in place
df.dropna(inplace=True)df.to_pickle('Before_Modeling_Grades')Note that loan status can be included in the features, and I do not consider it a data leakage program since in reality, we always have the loan status in hand. Our goal is to reproduce the grading system that Lending Club uses.
df.read_pickle('Before_Modeling_Grades')df.drop('sub_grade', axis =1)import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score, classification_report, confusion_matrix
import gcdef print_score(clf, X_train, y_train, X_test, y_test, train=True):
if train:
print("Train Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_train, clf.predict(X_train))))
print("Classification Report: \n {}\n".format(classification_report(y_train, clf.predict(X_train))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_train, clf.predict(X_train))))
res = cross_val_score(clf, X_train, y_train, cv=10, scoring='accuracy')
print("Average Accuracy: \t {0:.4f}".format(np.mean(res)))
print("Accuracy SD: \t\t {0:.4f}".format(np.std(res)))
elif train==False:
print("Test Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_test, clf.predict(X_test))))
print("Classification Report: \n {}\n".format(classification_report(y_test, clf.predict(X_test))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_test, clf.predict(X_test))))
from sklearn.model_selection import train_test_splitX_train, X_test, y_train, y_test = train_test_split(df.drop('grade',axis=1),df['grade'],test_size=0.15,random_state=101)gc.collect()8783
from sklearn.preprocessing import StandardScalersc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test=sc.transform(X_test)/opt/anaconda3/lib/python3.7/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype uint8, int64, float64 were all converted to float64 by StandardScaler.
return self.partial_fit(X, y)
/opt/anaconda3/lib/python3.7/site-packages/sklearn/base.py:462: DataConversionWarning: Data with input dtype uint8, int64, float64 were all converted to float64 by StandardScaler.
return self.fit(X, **fit_params).transform(X)
/opt/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:3: DataConversionWarning: Data with input dtype uint8, int64, float64 were all converted to float64 by StandardScaler.
This is separate from the ipykernel package so we can avoid doing imports until
from sklearn.ensemble import RandomForestClassifierclf_rf = RandomForestClassifier(n_estimators=10, random_state=21) # I changed it to 10 for faster training time
clf_rf.fit(X_train, y_train)RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=None,
oob_score=False, random_state=21, verbose=0, warm_start=False)
print_score(clf_rf, X_train, y_train, X_test, y_test, train=False)Test Result:
accuracy score: 0.7605
Classification Report:
precision recall f1-score support
0 0.69 0.88 0.77 94286
1 0.60 0.51 0.56 46374
2 0.82 0.86 0.84 94793
3 0.96 0.86 0.91 62402
4 0.49 0.22 0.30 19194
5 0.47 0.08 0.13 5807
6 0.42 0.01 0.02 1668
micro avg 0.76 0.76 0.76 324524
macro avg 0.64 0.49 0.50 324524
weighted avg 0.75 0.76 0.74 324524
Confusion Matrix:
[[83121 3432 7304 257 164 8 0]
[19081 23837 1443 60 1880 72 1]
[10969 349 81431 2032 12 0 0]
[ 585 37 8009 53771 0 0 0]
[ 5346 8971 426 10 4169 267 5]
[ 1276 2194 66 2 1801 449 19]
[ 304 594 30 1 556 165 18]]
!git clone --recursive https://github.com/microsoft/LightGBM
!cd LightGBM
!mkdir build
!cd build
!cmake -DUSE_GPU=1 ..Cloning into 'LightGBM'...
remote: Enumerating objects: 20, done.�[K
remote: Counting objects: 100% (20/20), done.�[K
remote: Compressing objects: 100% (17/17), done.�[K
remote: Total 13861 (delta 4), reused 8 (delta 3), pack-reused 13841�[K
Receiving objects: 100% (13861/13861), 9.69 MiB | 28.19 MiB/s, done.
Resolving deltas: 100% (9920/9920), done.
Submodule 'include/boost/compute' (https://github.com/boostorg/compute) registered for path 'compute'
Cloning into '/content/LightGBM/compute'...
remote: Enumerating objects: 72, done.
remote: Counting objects: 100% (72/72), done.
remote: Compressing objects: 100% (47/47), done.
remote: Total 21728 (delta 30), reused 44 (delta 19), pack-reused 21656
Receiving objects: 100% (21728/21728), 8.53 MiB | 30.03 MiB/s, done.
Resolving deltas: 100% (17550/17550), done.
Submodule path 'compute': checked out '36c89134d4013b2e5e45bc55656a18bd6141995a'
CMake Error: The source directory "/" does not appear to contain CMakeLists.txt.
Specify --help for usage, or press the help button on the CMake GUI.
?import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import roc_auc_score, roc_curve
from sklearn.model_selection import KFold, StratifiedKFold
from lightgbm import LGBMClassifier
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import gc
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score, classification_report, confusion_matrixdf=pd.read_pickle('Before_Modeling_Grades')
df = df.drop('sub_grade', axis =1)def kfold_lightgbm(train_df, num_folds, stratified = False):
print("Starting LightGBM. Train shape: {}".format(train_df.shape))
# Cross validation model
if stratified:
folds = StratifiedKFold(n_splits= num_folds, shuffle=True, random_state=47)
else:
folds = KFold(n_splits= num_folds, shuffle=True, random_state=47)
oof_preds = np.zeros(train_df.shape[0])
feature_importance_df = pd.DataFrame()
feats = [f for f in train_df.columns if f not in ['grade']]
# Splitting the training set into folds for Cross Validation
for n_fold, (train_idx, valid_idx) in enumerate(folds.split(train_df[feats], train_df['grade'])):
train_x, train_y = train_df[feats].iloc[train_idx], train_df['grade'].iloc[train_idx]
valid_x, valid_y = train_df[feats].iloc[valid_idx], train_df['grade'].iloc[valid_idx]
# LightGBM parameters found by Bayesian optimization
clf = LGBMClassifier(
nthread=12,#previous number 4
n_estimators=100, # Previous number 10000
learning_rate=0.1,
num_leaves=32,
colsample_bytree=0.9497036,
subsample=0.8715623,
max_depth=8,
reg_alpha=0.04,
reg_lambda=0.073,
min_split_gain=0.0222415,
min_child_weight=40,
silent=-1,
verbose=-1
#device= 'gpu',gpu_platform_id= 0,gpu_device_id= 0
)
# Fitting the model and evaluating by AUC
clf.fit(train_x, train_y, eval_set=[(train_x, train_y), (valid_x, valid_y)],
eval_metric= 'logloss', verbose= 1000, early_stopping_rounds= 200)
print_score(clf, train_x, train_y, valid_x, valid_y, train=False)
# Dataframe holding the different features and their importance
fold_importance_df = pd.DataFrame()
fold_importance_df["feature"] = feats
fold_importance_df["importance"] = clf.feature_importances_
fold_importance_df["fold"] = n_fold + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
# Freeing up memory
del clf, train_x, train_y, valid_x, valid_y
gc.collect()
display_importances(feature_importance_df)
return feature_importance_dfdef print_score(clf, X_train, y_train, X_test, y_test, train=True):
if train:
print("Train Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_train, clf.predict(X_train))))
print("Classification Report: \n {}\n".format(classification_report(y_train, clf.predict(X_train))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_train, clf.predict(X_train))))
res = cross_val_score(clf, X_train, y_train, cv=10, scoring='accuracy')
print("Average Accuracy: \t {0:.4f}".format(np.mean(res)))
print("Accuracy SD: \t\t {0:.4f}".format(np.std(res)))
elif train==False:
print("Test Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_test, clf.predict(X_test))))
print("Classification Report: \n {}\n".format(classification_report(y_test, clf.predict(X_test))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_test, clf.predict(X_test)))) def display_importances(feature_importance_df_):
cols = feature_importance_df_[["feature", "importance"]].groupby("feature").mean().sort_values(by="importance", ascending=False)[:40].index
best_features = feature_importance_df_.loc[feature_importance_df_.feature.isin(cols)]
plt.figure(figsize=(15, 12))
sns.barplot(x="importance", y="feature", data=best_features.sort_values(by="importance", ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
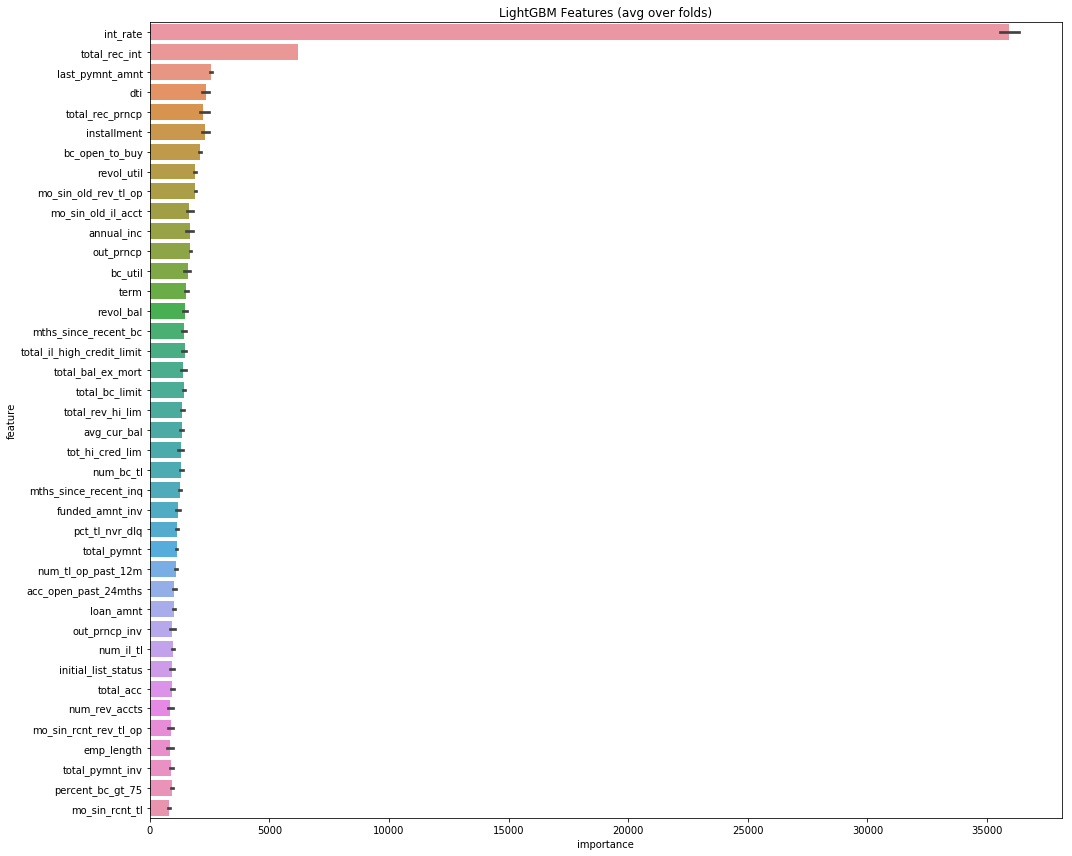
plt.savefig('lgbm_importances.png')feat_importance = kfold_lightgbm(df, num_folds= 3, stratified= False)Starting LightGBM. Train shape: (2163488, 1317)
Training until validation scores don't improve for 200 rounds.
[1000] training's multi_logloss: 0.0028125 training's multi_logloss: 0.0028125 valid_1's multi_logloss: 0.00508117 valid_1's multi_logloss: 0.00508117
Did not meet early stopping. Best iteration is:
[1000] training's multi_logloss: 0.0028125 training's multi_logloss: 0.0028125 valid_1's multi_logloss: 0.00508117 valid_1's multi_logloss: 0.00508117
Test Result:
accuracy score: 0.9986
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 209325
1 1.00 1.00 1.00 103212
2 1.00 1.00 1.00 211657
3 1.00 1.00 1.00 137587
4 0.99 1.00 1.00 42575
5 0.99 0.99 0.99 13062
6 0.98 0.96 0.97 3745
micro avg 1.00 1.00 1.00 721163
macro avg 0.99 0.99 0.99 721163
weighted avg 1.00 1.00 1.00 721163
Confusion Matrix:
[[209149 122 41 5 8 0 0]
[ 56 102993 4 2 157 0 0]
[ 104 3 211520 26 4 0 0]
[ 1 1 6 137579 0 0 0]
[ 7 99 0 1 42415 51 2]
[ 0 2 0 0 72 12910 78]
[ 0 0 0 0 7 135 3603]]
Training until validation scores don't improve for 200 rounds.
[1000] training's multi_logloss: 0.00263011 training's multi_logloss: 0.00263011 valid_1's multi_logloss: 0.00482896 valid_1's multi_logloss: 0.00482896
Did not meet early stopping. Best iteration is:
[1000] training's multi_logloss: 0.00263011 training's multi_logloss: 0.00263011 valid_1's multi_logloss: 0.00482896 valid_1's multi_logloss: 0.00482896
Test Result:
accuracy score: 0.9988
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 208644
1 1.00 1.00 1.00 103404
2 1.00 1.00 1.00 211804
3 1.00 1.00 1.00 137763
4 0.99 1.00 1.00 42858
5 0.99 0.99 0.99 12948
6 0.99 0.97 0.98 3742
micro avg 1.00 1.00 1.00 721163
macro avg 1.00 0.99 0.99 721163
weighted avg 1.00 1.00 1.00 721163
Confusion Matrix:
[[208507 84 43 5 5 0 0]
[ 30 103225 5 1 143 0 0]
[ 134 1 211654 14 1 0 0]
[ 3 0 9 137750 1 0 0]
[ 5 124 7 0 42665 51 6]
[ 2 4 0 0 70 12846 26]
[ 0 4 0 0 14 97 3627]]
Training until validation scores don't improve for 200 rounds.
[1000] training's multi_logloss: 0.00276678 training's multi_logloss: 0.00276678 valid_1's multi_logloss: 0.00515879 valid_1's multi_logloss: 0.00515879
Did not meet early stopping. Best iteration is:
[1000] training's multi_logloss: 0.00276678 training's multi_logloss: 0.00276678 valid_1's multi_logloss: 0.00515879 valid_1's multi_logloss: 0.00515879
Test Result:
accuracy score: 0.9986
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 209083
1 1.00 1.00 1.00 102822
2 1.00 1.00 1.00 211557
3 1.00 1.00 1.00 138042
4 0.99 1.00 1.00 42819
5 0.98 0.99 0.98 13073
6 0.97 0.96 0.96 3766
micro avg 1.00 1.00 1.00 721162
macro avg 0.99 0.99 0.99 721162
weighted avg 1.00 1.00 1.00 721162
Confusion Matrix:
[[208920 96 47 13 7 0 0]
[ 49 102612 3 3 155 0 0]
[ 94 1 211435 27 0 0 0]
[ 3 1 12 138026 0 0 0]
[ 4 105 5 1 42650 53 1]
[ 1 3 0 0 57 12884 128]
[ 1 2 0 0 12 152 3599]]
/opt/anaconda3/lib/python3.7/site-packages/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval
We use htop in command line to monitor the cpu usage. To our delight, LightGBM is able to multi thread since we set the number of threads to be 12, and our 12-core CPU were all used at full during training. However, the wait time is still very, very long
In the mean time for training, this is what I started doing
- Grabed some water, preferably sparkling water
- Started another jupyter notebook for subgrade modeling, with the knowledge that not all Google Cloud 12 Cores are being used. Run the bagging for subgrades.
- While waiting for (2). Started another instance trying to do lightGBM on a gpu. This is based on a docker file of lightGBM on gpu https://github.com/microsoft/LightGBM/tree/master/docker/gpu. Starting an PyTorch from NVIDIA Google Cloud instance with nv-docker from Google CloudThis is a new instance based on the docker file of LightGBM on GPU provided in the official github.
Side note: I encountered many problem installing the docker file, mostly a conda yes/no question, and I figured out a better way to do that. I ended up submitting a pull request to the LightGBM hosted by Microsoft. But even after the pull request, the docker is not set up properly as the preinstalled pandas version is too old to even read our pickle file. We spent 1-hour installing a new, clean environment with useable pandas, which also made the LightBGM-on-GPU not callable. We did not pursue further
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from fastai.tabular import *df = pd.read_pickle('0930SundayUntitled')Converting the datetime objects into regular objects
df['last_pymnt_d'] = df['last_pymnt_d'].dt.strftime('%Y-%m')
df['issue_d'] =df['issue_d'].dt.strftime('%Y-%m')
df['last_credit_pull_d']=df['last_credit_pull_d'].dt.strftime('%Y-%m')
df['earliest_cr_line']=df['earliest_cr_line'].dt.strftime('%Y-%m')Dealing with converting grade and subgrade into categories
factor_grade = pd.factorize(df['grade'])
df['grade'] = factor_grade[0]
factor_subgrade = pd.factorize(df['sub_grade'])
df['sub_grade'] = factor_subgrade[0]Dealing with term column
df['term'].replace(to_replace='[^0-9]+', value='', inplace=True, regex=True)
df['term']=df['term'].astype(int)Dealing with Employment Length
df['emp_length']= df['emp_length'].astype(int)#df.to_csv('BeforeFastai.csv')#df = pd.read_csv('BeforeFastai.csv')dep_var = 'grade'
df = df.drop('sub_grade',axis=1)cat_names = df.select_dtypes(include=object).columnsprocs = [FillMissing, Categorify, Normalize]
size=len(df)*0.2test_index = np.random.choice(df.index, size=int(len(df)*0.2), replace=False)cont_list, cat_list = cont_cat_split(df=df, max_card=25, dep_var='grade')cat_list['term',
'emp_length',
'home_ownership',
'verification_status',
'issue_d',
'pymnt_plan',
'purpose',
'addr_state',
'earliest_cr_line',
'initial_list_status',
'last_pymnt_d',
'last_credit_pull_d',
'policy_code',
'application_type',
'hardship_flag',
'disbursement_method',
'debt_settlement_flag',
'TARGET']
data = TabularDataBunch.from_df('BeforeFastai.csv', df, dep_var=dep_var, valid_idx=test_index, procs=procs, cat_names=cat_list)learn = tabular_learner(data, layers=[200,100], metrics=accuracy)learn.fit_one_cycle(1, 1e-2)| epoch | train_loss | valid_loss | accuracy | time |
|---|---|---|---|---|
| 0 | 0.234671 | 0.180922 | 0.979110 | 12:35 |
Notice that we achieved 0.979 accuracy with just one cycle trainig at 1e-2 learning rate.
Training time takes 12:35 minutes . This is a about 5 times faster than the most accurate model we tried, boosting with LightGBM, with bearable amount of performance compromise that should be able to minimized by further hyperparameter tuning.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from sklearn.metrics import roc_auc_score, roc_curve
from sklearn.model_selection import KFold, StratifiedKFold
from lightgbm import LGBMClassifier
import numpy as np
import gc
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.metrics import accuracy_score, classification_report, confusion_matrixdf = pd.read_pickle('Before_Modeling_Grades')df = df.drop('grade', axis =1)def print_score(clf, X_train, y_train, X_test, y_test, train=True):
if train:
print("Train Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_train, clf.predict(X_train))))
print("Classification Report: \n {}\n".format(classification_report(y_train, clf.predict(X_train))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_train, clf.predict(X_train))))
res = cross_val_score(clf, X_train, y_train, cv=10, scoring='accuracy')
print("Average Accuracy: \t {0:.4f}".format(np.mean(res)))
print("Accuracy SD: \t\t {0:.4f}".format(np.std(res)))
elif train==False:
print("Test Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_test, clf.predict(X_test))))
print("Classification Report: \n {}\n".format(classification_report(y_test, clf.predict(X_test))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_test, clf.predict(X_test)))) from sklearn.model_selection import train_test_splitX_train, X_test, y_train, y_test = train_test_split(df.drop('sub_grade',axis=1),df['sub_grade'],test_size=0.15,random_state=101)
gc.collect()57
from sklearn.preprocessing import StandardScalersc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test=sc.transform(X_test)/home/alexanderli/anaconda3/lib/python3.7/site-packages/sklearn/preprocessing/data.py:645: DataConversionWarning: Data with input dtype uint8, int64, float64 were all converted to float64 by StandardScaler.
return self.partial_fit(X, y)
/home/alexanderli/anaconda3/lib/python3.7/site-packages/sklearn/base.py:464: DataConversionWarning: Data with input dtype uint8, int64, float64 were all converted to float64 by StandardScaler.
return self.fit(X, **fit_params).transform(X)
/home/alexanderli/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:3: DataConversionWarning: Data with input dtype uint8, int64, float64 were all converted to float64 by StandardScaler.
This is separate from the ipykernel package so we can avoid doing imports until
from sklearn.ensemble import RandomForestClassifierclf_rf = RandomForestClassifier(n_estimators=10, random_state=21)
clf_rf.fit(X_train, y_train)
print_score(clf_rf, X_train, y_train, X_test, y_test, train=False)Test Result:
accuracy score: 0.2473
Classification Report:
precision recall f1-score support
0 0.21 0.41 0.28 21033
1 0.12 0.19 0.15 10328
2 0.14 0.18 0.16 11777
3 0.20 0.29 0.24 18524
4 0.20 0.24 0.22 18653
5 0.19 0.20 0.19 19187
6 0.12 0.09 0.10 6906
7 0.23 0.29 0.26 18524
8 0.28 0.37 0.32 13608
9 0.29 0.28 0.29 19849
10 0.21 0.18 0.19 16889
11 0.12 0.07 0.09 8162
12 0.10 0.05 0.07 4684
13 0.10 0.04 0.05 3242
14 0.29 0.25 0.27 20010
15 0.13 0.07 0.09 9201
16 0.70 0.84 0.76 12757
17 0.11 0.04 0.06 3182
18 0.27 0.22 0.24 18411
19 0.30 0.24 0.27 17999
20 0.32 0.23 0.27 15385
21 0.04 0.01 0.01 711
22 0.32 0.19 0.24 10489
23 0.10 0.03 0.05 3869
24 0.44 0.25 0.31 10163
25 0.10 0.03 0.05 4217
26 0.09 0.01 0.02 881
27 0.05 0.01 0.01 559
28 0.06 0.01 0.01 376
29 0.09 0.02 0.03 1929
30 0.05 0.01 0.01 1249
31 0.07 0.01 0.02 1037
32 0.08 0.00 0.01 213
33 0.00 0.00 0.00 310
34 0.00 0.00 0.00 210
micro avg 0.25 0.25 0.25 324524
macro avg 0.17 0.15 0.15 324524
weighted avg 0.24 0.25 0.24 324524
Confusion Matrix:
[[8577 289 377 ... 0 0 0]
[1038 1916 1525 ... 0 0 0]
[1196 1599 2163 ... 0 0 0]
...
[ 6 25 17 ... 1 0 0]
[ 13 38 25 ... 0 0 0]
[ 9 27 16 ... 1 0 0]]
We are not very happy with the 0.24 accuracy. Let us proceed to boosting with LightGBM for subgrade classification
def kfold_lightgbm(train_df, num_folds, stratified = False):
print("Starting LightGBM. Train shape: {}".format(train_df.shape))
# Cross validation model
if stratified:
folds = StratifiedKFold(n_splits= num_folds, shuffle=True, random_state=47)
else:
folds = KFold(n_splits= num_folds, shuffle=True, random_state=47)
oof_preds = np.zeros(train_df.shape[0])
feature_importance_df = pd.DataFrame()
feats = [f for f in train_df.columns if f not in ['sub_grade']]
# Splitting the training set into folds for Cross Validation
for n_fold, (train_idx, valid_idx) in enumerate(folds.split(train_df[feats], train_df['sub_grade'])):
train_x, train_y = train_df[feats].iloc[train_idx], train_df['sub_grade'].iloc[train_idx]
valid_x, valid_y = train_df[feats].iloc[valid_idx], train_df['sub_grade'].iloc[valid_idx]
# LightGBM parameters found by Bayesian optimization
clf = LGBMClassifier(
nthread=12,#previous number 4
n_estimators=1000, # Previous number 10000
learning_rate=0.02,
num_leaves=32,
colsample_bytree=0.9497036,
subsample=0.8715623,
max_depth=8,
reg_alpha=0.04,
reg_lambda=0.073,
min_split_gain=0.0222415,
min_child_weight=40,
silent=-1,
verbose=-1
#device_type=gpu,gpu_platform_id= 0,gpu_device_id= 0
)
# Fitting the model and evaluating by AUC
clf.fit(train_x, train_y, eval_set=[(train_x, train_y), (valid_x, valid_y)],
eval_metric= 'logloss', verbose= 1000, early_stopping_rounds= 200)
print_score(clf, train_x, train_y, valid_x, valid_y, train=False)
# Dataframe holding the different features and their importance
fold_importance_df = pd.DataFrame()
fold_importance_df["feature"] = feats
fold_importance_df["importance"] = clf.feature_importances_
fold_importance_df["fold"] = n_fold + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
# Freeing up memory
del clf, train_x, train_y, valid_x, valid_y
display_importances(feature_importance_df)
return feature_importance_dfdef print_score(clf, X_train, y_train, X_test, y_test, train=True):
if train:
print("Train Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_train, clf.predict(X_train))))
print("Classification Report: \n {}\n".format(classification_report(y_train, clf.predict(X_train))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_train, clf.predict(X_train))))
res = cross_val_score(clf, X_train, y_train, cv=10, scoring='accuracy')
print("Average Accuracy: \t {0:.4f}".format(np.mean(res)))
print("Accuracy SD: \t\t {0:.4f}".format(np.std(res)))
elif train==False:
print("Test Result:\n")
print("accuracy score: {0:.4f}\n".format(accuracy_score(y_test, clf.predict(X_test))))
print("Classification Report: \n {}\n".format(classification_report(y_test, clf.predict(X_test))))
print("Confusion Matrix: \n {}\n".format(confusion_matrix(y_test, clf.predict(X_test)))) def display_importances(feature_importance_df_):
cols = feature_importance_df_[["feature", "importance"]].groupby("feature").mean().sort_values(by="importance", ascending=False)[:40].index
best_features = feature_importance_df_.loc[feature_importance_df_.feature.isin(cols)]
plt.figure(figsize=(15, 12))
sns.barplot(x="importance", y="feature", data=best_features.sort_values(by="importance", ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
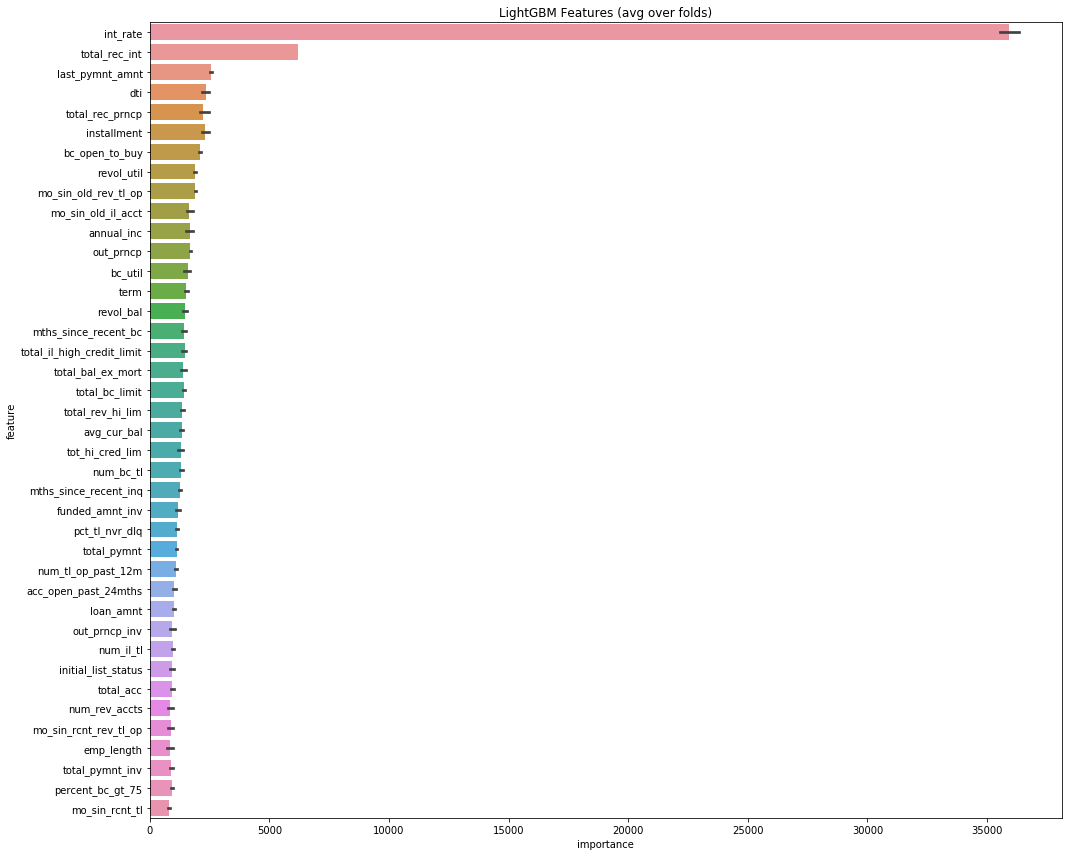
plt.savefig('lgbm_importances.png')feat_importance = kfold_lightgbm(df, num_folds= 3, stratified= False)Starting LightGBM. Train shape: (2163488, 1317)
Training until validation scores don't improve for 200 rounds.
[1000] training's multi_logloss: 0.0028125 training's multi_logloss: 0.0028125 valid_1's multi_logloss: 0.00508117 valid_1's multi_logloss: 0.00508117
Did not meet early stopping. Best iteration is:
[1000] training's multi_logloss: 0.0028125 training's multi_logloss: 0.0028125 valid_1's multi_logloss: 0.00508117 valid_1's multi_logloss: 0.00508117
Test Result:
accuracy score: 0.9986
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 209325
1 1.00 1.00 1.00 103212
2 1.00 1.00 1.00 211657
3 1.00 1.00 1.00 137587
4 0.99 1.00 1.00 42575
5 0.99 0.99 0.99 13062
6 0.98 0.96 0.97 3745
micro avg 1.00 1.00 1.00 721163
macro avg 0.99 0.99 0.99 721163
weighted avg 1.00 1.00 1.00 721163
Confusion Matrix:
[[209149 122 41 5 8 0 0]
[ 56 102993 4 2 157 0 0]
[ 104 3 211520 26 4 0 0]
[ 1 1 6 137579 0 0 0]
[ 7 99 0 1 42415 51 2]
[ 0 2 0 0 72 12910 78]
[ 0 0 0 0 7 135 3603]]
Training until validation scores don't improve for 200 rounds.
[1000] training's multi_logloss: 0.00263011 training's multi_logloss: 0.00263011 valid_1's multi_logloss: 0.00482896 valid_1's multi_logloss: 0.00482896
Did not meet early stopping. Best iteration is:
[1000] training's multi_logloss: 0.00263011 training's multi_logloss: 0.00263011 valid_1's multi_logloss: 0.00482896 valid_1's multi_logloss: 0.00482896
Test Result:
accuracy score: 0.9988
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 208644
1 1.00 1.00 1.00 103404
2 1.00 1.00 1.00 211804
3 1.00 1.00 1.00 137763
4 0.99 1.00 1.00 42858
5 0.99 0.99 0.99 12948
6 0.99 0.97 0.98 3742
micro avg 1.00 1.00 1.00 721163
macro avg 1.00 0.99 0.99 721163
weighted avg 1.00 1.00 1.00 721163
Confusion Matrix:
[[208507 84 43 5 5 0 0]
[ 30 103225 5 1 143 0 0]
[ 134 1 211654 14 1 0 0]
[ 3 0 9 137750 1 0 0]
[ 5 124 7 0 42665 51 6]
[ 2 4 0 0 70 12846 26]
[ 0 4 0 0 14 97 3627]]
Training until validation scores don't improve for 200 rounds.
[1000] training's multi_logloss: 0.00276678 training's multi_logloss: 0.00276678 valid_1's multi_logloss: 0.00515879 valid_1's multi_logloss: 0.00515879
Did not meet early stopping. Best iteration is:
[1000] training's multi_logloss: 0.00276678 training's multi_logloss: 0.00276678 valid_1's multi_logloss: 0.00515879 valid_1's multi_logloss: 0.00515879
Test Result:
accuracy score: 0.9986
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 209083
1 1.00 1.00 1.00 102822
2 1.00 1.00 1.00 211557
3 1.00 1.00 1.00 138042
4 0.99 1.00 1.00 42819
5 0.98 0.99 0.98 13073
6 0.97 0.96 0.96 3766
micro avg 1.00 1.00 1.00 721162
macro avg 0.99 0.99 0.99 721162
weighted avg 1.00 1.00 1.00 721162
Confusion Matrix:
[[208920 96 47 13 7 0 0]
[ 49 102612 3 3 155 0 0]
[ 94 1 211435 27 0 0 0]
[ 3 1 12 138026 0 0 0]
[ 4 105 5 1 42650 53 1]
[ 1 3 0 0 57 12884 128]
[ 1 2 0 0 12 152 3599]]
/opt/anaconda3/lib/python3.7/site-packages/scipy/stats/stats.py:1713: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
return np.add.reduce(sorted[indexer] * weights, axis=axis) / sumval
For a training time of 25 minutes and 25 secs, boosting training is finished with 0.9986 accuracy
Importing the data
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from fastai.tabular import *df = pd.read_pickle('0930SundayUntitled')Converting the datetime objects into regular objects
df['last_pymnt_d'] = df['last_pymnt_d'].dt.strftime('%Y-%m')
df['issue_d'] =df['issue_d'].dt.strftime('%Y-%m')
df['last_credit_pull_d']=df['last_credit_pull_d'].dt.strftime('%Y-%m')
df['earliest_cr_line']=df['earliest_cr_line'].dt.strftime('%Y-%m')Dealing with converting grade and subgrade into categories
factor_grade = pd.factorize(df['grade'])
df['grade'] = factor_grade[0]
factor_subgrade = pd.factorize(df['sub_grade'])
df['sub_grade'] = factor_subgrade[0]Dealing with term column
df['term'].replace(to_replace='[^0-9]+', value='', inplace=True, regex=True)
df['term']=df['term'].astype(int)Dealing with Employment Length
df['emp_length']= df['emp_length'].astype(int)df.to_csv('BeforeFastai.csv')df = pd.read_csv('BeforeFastai.csv')dep_var = 'sub_grade'
df = df.drop('grade',axis=1)cat_names = df.select_dtypes(include=object).columnsprocs = [FillMissing, Categorify, Normalize]
size=len(df)*0.2test_index = np.random.choice(df.index, size=int(len(df)*0.2), replace=False)cont_list, cat_list = cont_cat_split(df=df, max_card=25, dep_var='sub_grade')cat_list['term',
'emp_length',
'home_ownership',
'verification_status',
'issue_d',
'pymnt_plan',
'purpose',
'addr_state',
'earliest_cr_line',
'initial_list_status',
'last_pymnt_d',
'last_credit_pull_d',
'policy_code',
'application_type',
'hardship_flag',
'disbursement_method',
'debt_settlement_flag',
'TARGET']
data = TabularDataBunch.from_df('BeforeFastai.csv', df, dep_var=dep_var, valid_idx=test_index, procs=procs, cat_names=cat_list)learn = tabular_learner(data, layers=[200,100], metrics=accuracy)learn.fit_one_cycle(1, 1e-2)| epoch | train_loss | valid_loss | accuracy | time |
|---|---|---|---|---|
| 0 | 1.162667 | 103.812439 | 0.877249 | 12:54 |
We are very happy with being able to achieve 0.877249 accuracy, with just 12:54 minutes of time in fastai. Compared to 0.9939 with boosting and 0.24 with bagging(random forrest), fast ai achieves accpetable amount of accuracy with less than one tenth training than than boosting.
In both cases of modeling grading and subgrading, we see that the fast.ai neural nets are the best model in terms of best balance between training time and performance. In production environment, we might still use LightGBM and boosting for the ultimate benchmark. But during traiing, it is both too long and distracting to use any training method that simply takes more than a cup-of-coffee time.

- why cup-of-coffee time is important?
If it takes more than what is needed to consume a cup of coffee, our data scientists would most likely be distracted during training. He/She might be browsing Hackernews, going to social media, engage in small talks, and he/she will most likely forget what hyperparameters were used along the way. As a enginner, we really want the flow to stay productive.
**update We were able to shrink down the training time by hyperparameter tuning, from over three hour to 25 minutes. **
##LightGBM issues Microsoft has responded to my pull request regarding the LightBGM docker stuck in the permission granting phase during the installation here. From our conversation, I think they are considering some updates. I tried to follow their guide to install it from Github instead(which was a pain given the lack of docs and quality of writing).
There are some problems with LightBGM support for GPU:
-
The three parameters, device, gpu_platform_id and gpu_device_id was a bit confusing for beginners.
-
You can only get GPU support by installing lightbgm from the offical github repo, compile it from CMAKE, and go to their subdirectory 'python-package' to install the python interface with 'install-gpu' specifically requested. Again confusing. Most people would need to uninstall their lightbgm from pip/conda, and do this again manually.
-
The LightBGMclassifer does not have support for GPU.
I would like to take this further by potentially developing a high level wrapper of LightGBM on GPU like how FastAI has native support.
This is my individual work, with the help of Interent/Stackoverflow/Kaggle Kernels. I was inspired by the kernals listed by Kaggle Lending Club dataset, and made significant changes to my logic flow and use case
Alex Shengzhi Li, 2019. alex@alexli.me