- 本仓库由医工科研团队kim老师维护
- 本仓库从rhhao/csMR拉取分支 在这个仓库的基础上 进一步增加了下方的功能
- 完善window平台的流程
- 完善各种环境的配置方法
- 增加了数据的格式化方法
- 扩展了更多的qtl数据
- window部分 R环境升级成4.4.1 TwoSampleMR包升级到0.6.8
- 增加部分的所有权归属医工科研团队所有
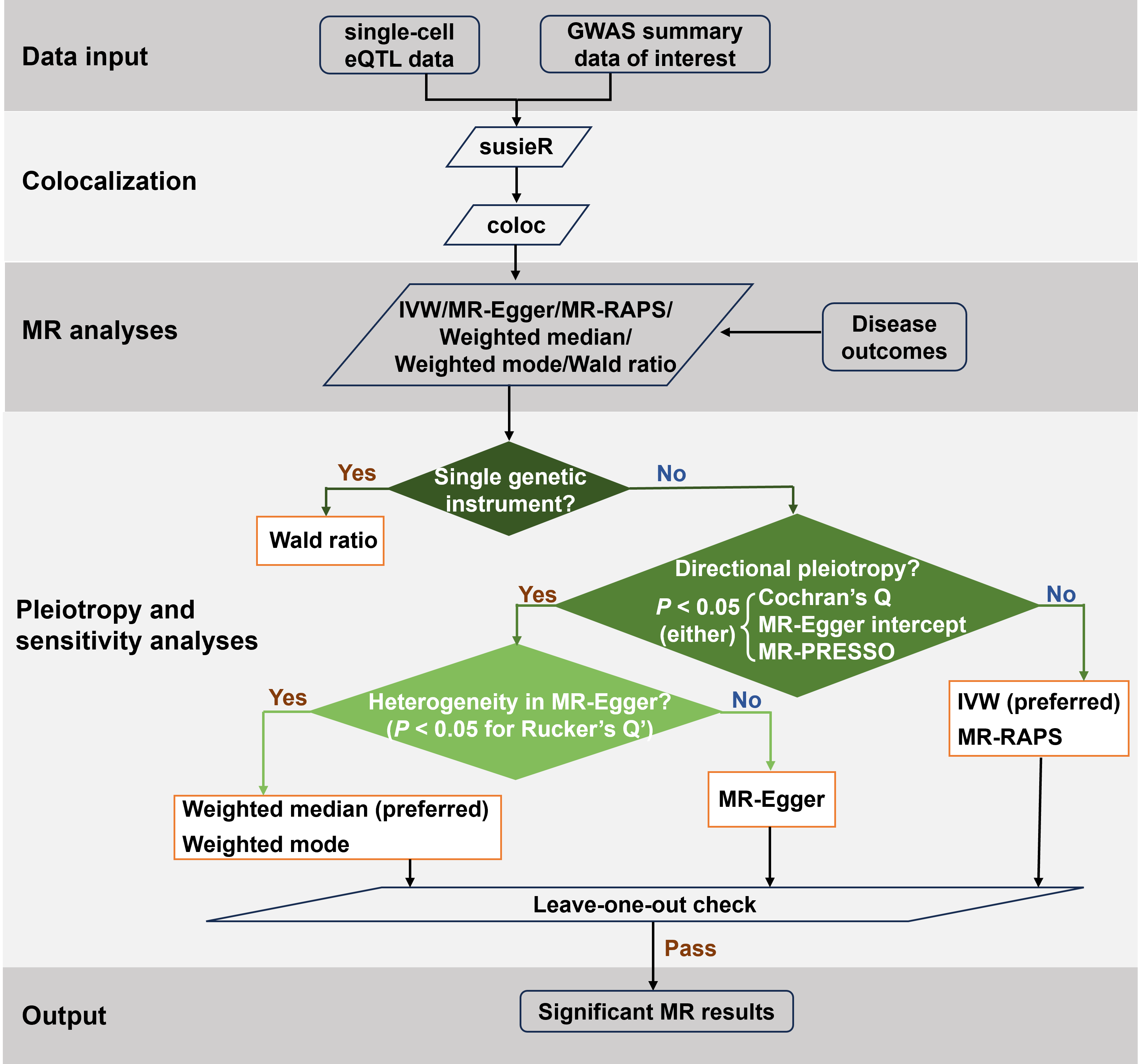
We introduce the Cell-Stratified Mendelian Randomization (csMR) framework to find cell-stratified causality of complex traits/diseases by integrating GWAS and single-cell eQTL data.
csMR can be downloaded by cloning this repository via the commands:
git clone https://github.com/rhhao/csMR.git
cd csMR In order to void any issues with software versioning, csMR utilises conda environments to automatically install all necessary dependencies. If conda is not present on your system, please install anaconda or miniconda following the official instructions.
Once conda is ready, users can create an environment to install dependencies that csMR needs though the following commands:
conda install -c conda-forge mamba
mamba env create --file envs/envpy3.yml
conda activate csMRcsMR requires R packages phenoscanner and updated mr.raps (version 0.4.1). To install these libraries, run the following commands:
if (!require("devtools")) { install.packages("devtools") }
devtools::install_github("phenoscanner/phenoscanner", force = TRUE, lib="<your installation path to conda>/envs/csMR/lib/R/library")
devtools::install_github("qingyuanzhao/mr.raps", force = TRUE, lib="<your installation path to conda>/envs/csMR/lib/R/library") Notice: in order to successfully run csMR, we suggest to set your temporary R library paths byexport R_LIBS=<your installation path to conda>/envs/csMR/lib/R/library:$R_LIBS before running csMR.
csMR requires PLINK (version 1.90) to calculate LD matrix. Follow the official instructions to install PLINK: https://www.cog-genomics.org/plink2/.
Step 1: Data preparation
csMR requires summary-level GWAS and QTL data that consist of at least 8 required columns for GWAS and 9 required columns for QTL. Other columns will be ignored during processing.
Notice: the names of required columns MUST be consistent with the following headers, while the order can be inconsistent. The required data format is presented as follows:
GWAS summary stats:
| SNP | A1 | A2 | MAF | BETA | SE | P | N |
|---|---|---|---|---|---|---|---|
| rs1 | A | T | 0.125 | 0.166 | 0.178 | 0.734 | 480000 |
| … | … | … | … | … | … | … | … |
| rs256 | C | G | 0.201 | 0.037 | 0.079 | 0.349 | 480000 |
QTL summary stats:
| SNP | GENE | A1 | A2 | MAF | BETA | SE | P | N |
|---|---|---|---|---|---|---|---|---|
| rs1 | geneY | A | T | 0.359 | -0.039 | 0.094 | 0.082 | 215 |
| … | … | … | … | … | … | … | … | … |
| rs256 | geneX | C | G | 0.229 | 0.064 | 0.012 | 0.779 | 215 |
Step 2: Modify the config.yml file
Before running csMR, a configuration file needs to be modified to specify the input data and advanced parameters. An example is provided in the config.yml file that will process the example data obtained by further download. The demo data are ready to use after decompressing into the installation directory of csMR.
Reference genome data are also required to run example data. Users can download reference_genome_1000G_EUR here. The genome data are ready to use after decompressing into the csMR/data directory.
Step 3: Run csMR
Now you have everything csMR needs, then you can run the analysis workflow under your installation directory by:
snakemake -s work_flow.snakefile --configfile config.yml -jWe recommend running with -j as it will use all available cores. Specifying -j 4 will use up to 4 cores.
Step 4: Inspect the output
The results of colocalization and MR will be placed in <your output dir>/COLCO and <your output dir>/MR respectively.
Ruo-Han Hao (Xi’an Jiaotong University, China)
Feng Jiang (Xi’an Jiaotong University, China)
Tian-Pei Zhang (Xi’an Jiaotong University, China)
Jun-Hui Liu (Xi’an Jiaotong University, China)
Please create an issue on the github repo if you encounter any problems. You can also contact the developers through email: ruohanhao@xjtu.edu.cn.
Cell-Stratified MR (csMR) Hao, RH., Zhang, TP., Jiang, F. et al. Revealing brain cell-stratified causality through dissecting causal variants according to their cell-type-specific effects on gene expression. Nat Commun 15, 4890 (2024). https://doi.org/10.1038/s41467-024-49263-4