FlexEMG_V2: A wearable biosensing system with printed flexible electrodes and in-sensor adaptive learning
This repository provides the offline dataset and MATLAB scripts used to verify the data acquision functionality of our device and the utilized algorithm. It is publicly available under GNU General Public License v3.
System requirements
- MATLAB (MathWorks, Inc.). Tested on version R2019a.
Installation guide
Install MATLAB, add the whole flexemg_v2/ directory to the path, and you are ready to go!
Repo structure
Note that all scripts are commented with function descriptions, input arguments, returns, etc.
dataset/: Dataset containing raw EMG signals from 5 subjects and 21 hand gestures with 5 repeatitions of each.functions/: MATLAB functions to emulate the HD algorithm being run on the on-board FPGA.
Filename structure
The files are named as "sss_e_ggg_t.mat" where sss is the subject number, e is the experiment number, ggg is the gesture ID, and t is the trail number.
The list of experimetns is as follows:
| Experiments | Gestures | Context |
|---|---|---|
| 1 | Single-DOF | Baseline, relaxed arm position |
| 2 | Multi-DOF | Baseline, relaxed arm position |
| 3 | Single-DOF | New arm position (arm wrestle) |
| 4 | Effort level | Low effort, relaxed arm position |
| 5 | Effort level | Medium effort, relaxed arm position |
| 6 | Effort level | High effort, relaxed arm position |
| 7 | Single-DOF | New wear session, relaxed arm position |
| 8 | Single-DOF | Prolonged wear, relaxed arm position |
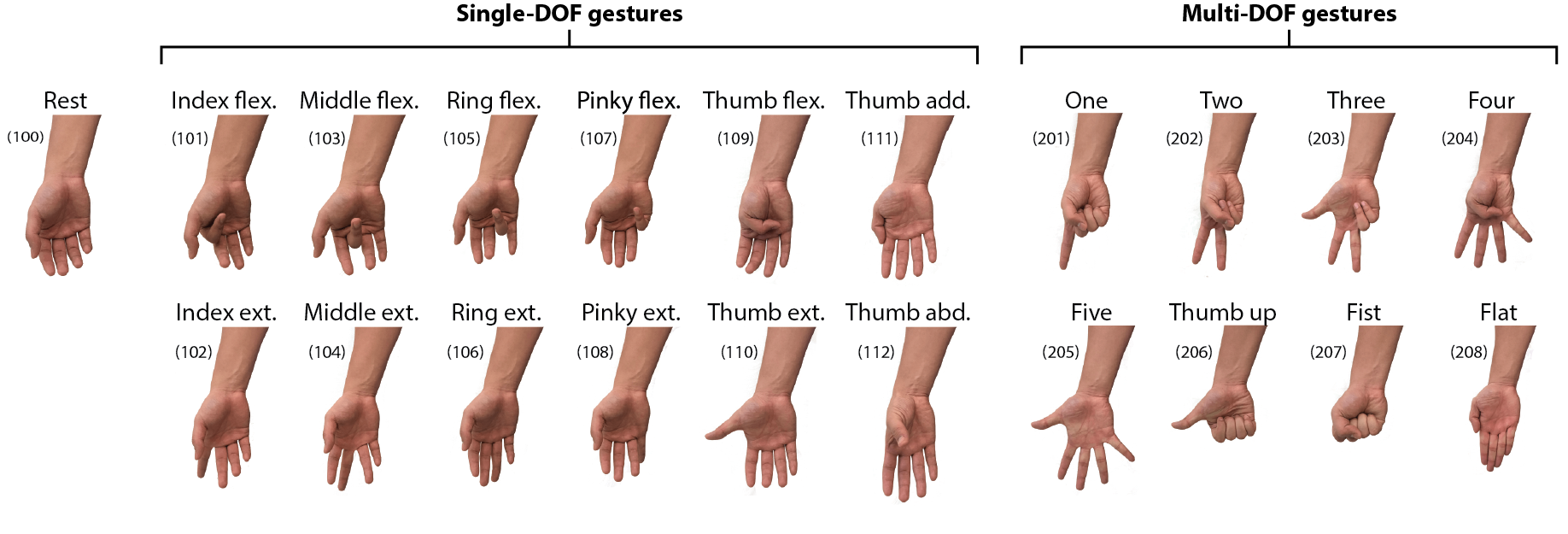
The list of gesture IDs is listed here:
Sample usage
To find the classification accuracy for subject 1 without using parallel computing toolbox, all you need to do is running all_expermients function in the following way:
all_expermients(1, 0)
The results will be saved in output/ folder with classification accuracy results for each run.
Problems?
If you face any problems or discover any bugs, please let us know: MyLastName AT berkeley DOT edu