The Human Immunology Project Consortium (HIPC) Dashboard is a web-application supporting the storage and dissemination of immune signatures curated from published literature. This GitHub repository is used for preparing data releases for the HIPC Dashboard. It includes both the actual release files as well as the code used for generating them. The HIPC Dashboard web-app itself is managed via a separate GitHub repository.
Our main objective in this project is to standardize the representation of immune signatures, i.e., of research findings that describe how immune exposures (e.g., infection, immunization) modulate the abundance of biologically important response components (e.g., genes, cell subpopulations). Such findings are typically not presented in a consistent format and may be found as text, tables, or images within publications. To improve access and to promote reuse, we have developed a data model that standardizes the content and context of published immune signatures. The model is used as the basis for generating curation spreadsheets which are utilized by human experts to record immune signatures from published literature. These spreadsheets are then further processed by the pipeline in this GitHub repository. The pipeline performs quality control, enforces consistent use of controlled vocabularies, and, finally, generates the data release files which will be uploaded to the HIPC Dashboard.
The repository contents are as follows:
-
./code: contains the R code used for processing the source curation data and generating the Dashboard release files.
-
./data:
-
./data/source_curations: source curation spreadsheets, as provided by curators.
-
./data/standardized_curations: curation spreadsheets in denormalized form, after undergoing processing for quality control and standardization. This is the state from which all other files are created, i.e. the convenience files and release files.
-
./data/reference_files: resources used to support the standardization process, including controlled vocabularies from external sources and mapping files maintained by the project.
-
./data/release_files: Dashboard release files. These are transformed, partially denormalized spreadsheet files generated from the standardized curations. They are formatted as needed for uploading to the Dashboard.
-
./data/convenience_files: the standardized curations conveniently reformatted to support human inspection and downstream computational analysis.
-
-
./docs: curation templates and column specification.
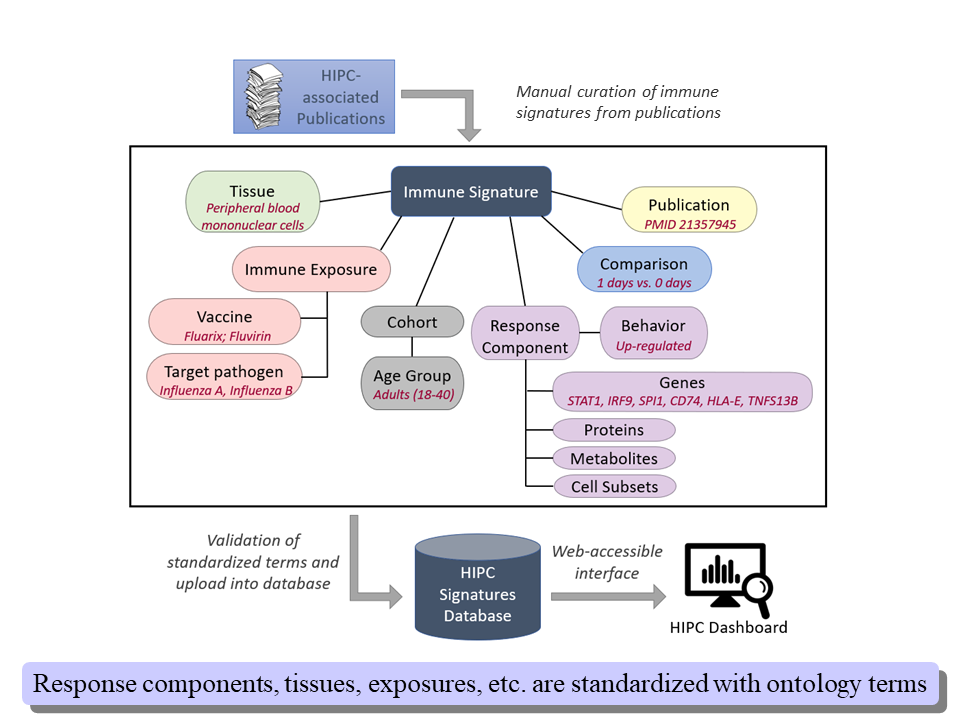
The overall workflow of the project is depicted below.
The figure above outlines the key elements of the immune signature data model. These are:
-
Immune exposure: the immunological perturbation whose effect is measured by the immune signature.
-
Response component: the biological entity whose abundance is being quantified.
-
Tissue: the biological context where the abundance measurements are made.
-
Cohort: the group of individuals subjected to the immune exposure; they are the source of the biological material from which the abundance measurements are taken.
-
Comparison: the conditions under which differential abundance is assessed.
For each of these elements we define a number of associated data fields used to collect element-specific information. As much as possible, values for these fields come from controlled vocabularies, either leveraging community standards (HUGO gene symbols for genes, Cell Ontology terms for cell types, Vaccine Ontology ids for vaccines, etc.) or utilizing term lists developed specifically for the needs of the project. A specification of all available data fields can be found under the ./docs directory.
Curators review selected publications, identify immune signatures reported therein, and record the signature-related data into the curation templates. Data is recorded as it appears in a publication and then standardized using suitable controlled vocabularies. The standardization is performed partly by the curators and partly by post-curation script-assisted annotation. For some data (tissue types, vaccines, pathogens) curators are asked to match what is reported in the publication to standardized terms from specific ontologies. For other data, such as gene symbols, the post-curation process will ensure that these are valid HUGO symbols and rectify issues such as spelling errors and use of deprecated terms. In all cases, the original text from the publication is also preserved, to be used for quality control and for maintaining proper provenance.
Dedicated templates are used for different types of immune signatures (infection, vaccination) to accommodate the specific data collection needs of each signature type. Blank copies ave available under the ./docs directory.
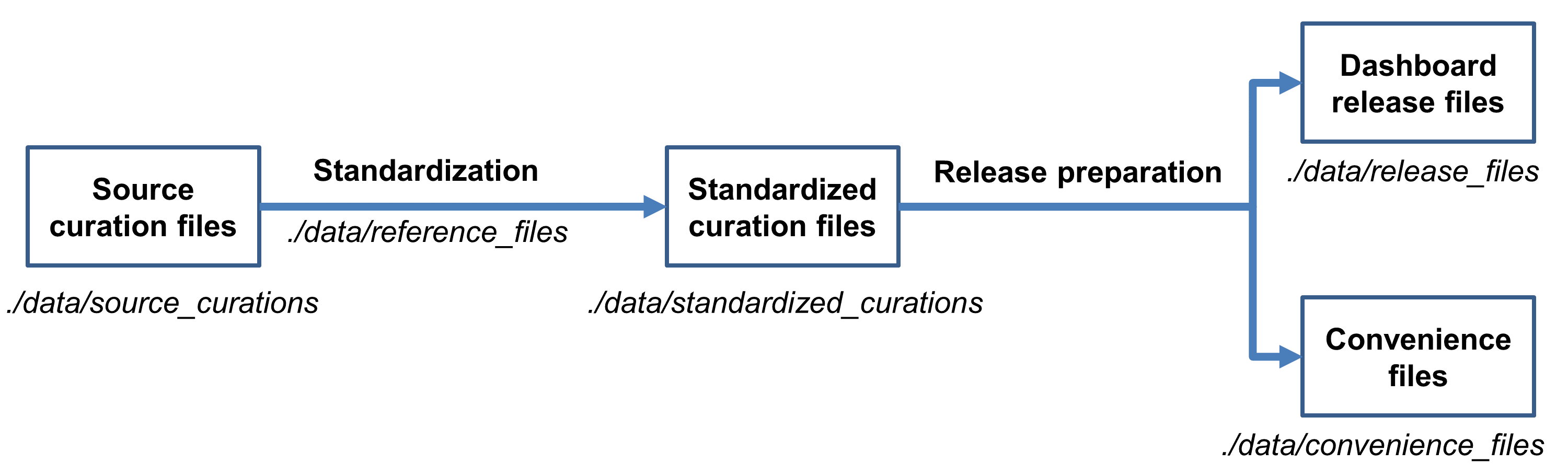
The high-level post-curation data processing workflow is outlined in the figure below.
Unprocessed data from the curation templates are stored as tab-delimited spreadsheet files in the directory ./data/source_curations. In preparation for data release, the files undergo a quality control and standardization process. The objective of the process is to identify and correct discrepancies and to ensure consistent use of controlled vocabularies. Some of the steps undertaken during this process include:
-
Gene symbols - Curated gene symbols are updated to current HGNC/NCBI symbols based on (1) NCBI synonyms and (2) a manually created mapping table. The later, where possible, resolves problematic symbols found in the curated data to their correct gene symbol, following manual re-examination of the original publication.
-
Cell types and markers - Measurement of cell type abundance typically involves flow cytometry experiments that utilize cell-marker based sorting. In these cases, Cell Ontology alone may not provide adequate resolution for the proper description of the cell types reported in an immune signature. To address this issue, we curate a mapping table to standardize the original publication cell type descriptions using terms from the Cell Ontology for cell types and Protein Ontology for additional type-defining markers.
-
Vaccines – For vaccination signatures, curators use the Vaccine Ontology to code vaccines and the NCBI Taxonomy to code the pathogens targeted by these vaccines. For influenza (and possibly other vaccines in the future) we remove the coding burden from the curators by utilizing a mapping file to perform this task. Curators only need to specify the vaccine using a year-based tag and then the coding information is retrieved by looking up the tag in the mapping file.
The curated files mentioned above as well as ancillary files utilized by the standardization process (e.g., containing controlled vocabulary terms from outside ontologies) are stored in ./data/reference_files and are updated with the latest versions prior to a Dashboard release.
The quality control and standardization process produces new, denormalized versions of the source spreadsheets which are placed under ./data/standardized_curations. These files then undergo further processing to generate the final data release files which will be uploaded to the Dashboard. This processing is essentially a straightforward repackaging of the spreadsheets into a format appropriate for the upload scripts. It involves splitting immune signatures into individual spreadsheet files, one signature per file. The final release files are stored under ./data/release_files and have the same columns as the standardized curation files, with some additions to support the requirements of the Dashboard itself and to preserve original curated values for fields updated by the pipeline.
The release preparation process also generates a number of convenience files, i.e., partially re-normalized versions of the standardized curations. These files are available in spreadsheet format, to facilitate human inspection; and in the Broad GMT tab delimited file format, to support downstream computational processing. The files are stored under ./data/convenience_files
Additional details about the processing pipeline can be found under the ./data directory.
The Dashboard database is reloaded in its entirety each time a new release is created. This involves reprocessing all curation templates, old and new, as described above. When the data processing is complete, the entire repository is tagged with the new Dashboard release number. This includes the pipeline code used to generate the release, the source curation templates, all intermediate and support files, and the final data release files.
The file changelog.txt in the root directory lists all major changes to the pipeline code and process in each release. The file changelog-data.txt in the root directory lists all major updates to the curation templates and the data in each release.
This project is licensed under the BSD 3-clause license. See the LICENSE.txt file for details.