This repo contains a simple code on a BERT-based dependency parser. I wanted to build one from scratch and ended up being surprised by how easy it has become at the current level of NLP.
The main inspiration is Dozat and Manning as well as Kondratyuk and Straka.
Please install git-lfs before installing
git clone https://github.com/fractalego/tree_parser.git
cd tree_parser
virtualenv .env --python=/usr/bin/python3
pip install .An example code is as below (in the file predict.py)
import os
from networkx.drawing.nx_agraph import write_dot
from tree_parser.parser import DependencyParser
_path = os.path.dirname(__file__)
_save_filename = os.path.join(_path, '../data/tree_parser.model')
_text = """
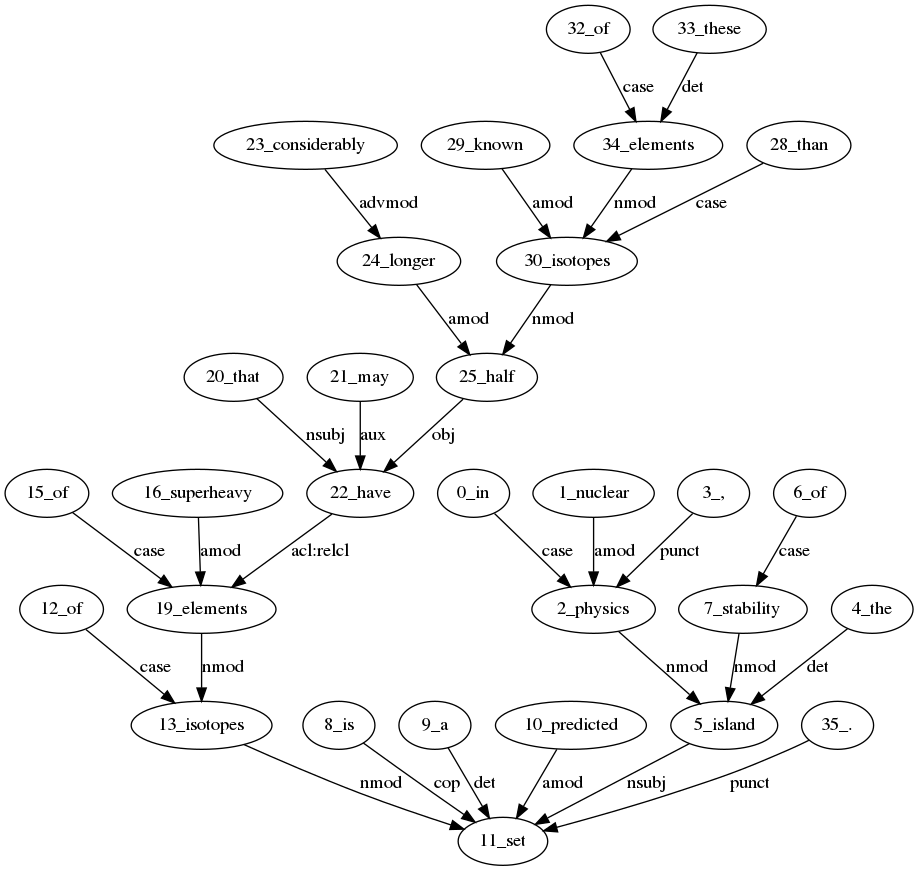
In nuclear physics, the island of stability is a predicted set of isotopes of superheavy elements
that may have considerably longer half-lives than known isotopes of these elements.
"""
if __name__ == '__main__':
parser = DependencyParser(_save_filename)
g = parser.parse(_text)
write_dot(g, 'test.dot')Which parses the sentence
In nuclear physics, the island of stability is a predicted set of isotopes of superheavy elements that may have considerably longer half-lives than known isotopes of these elements.
And outputs a '.dot' file.
The file can be converted onto a png using graphviz
dot -Tpng test.dot -o foo.pngThe output of DependencyParser().parse() is a Networkx graph.
Each node has two attributes:
- 'pos', the POS tag
- 'token', the parsed word.
Each edge has only one attribute 'label'. They can be accessed through the
In addition the ID associated to each node is of the form <INDEX>_TOKEN
In the previous example the code
import networkx as nx
print('Node words:')
print(nx.get_node_attributes(g, 'token'))
print('Node POS tags:')
print(nx.get_node_attributes(g, 'pos'))
print('edge labels:')
print(nx.get_edge_attributes(g, 'label'))would output
Node words:
{'0_in': 'in', '2_physics': 'physics', '1_nuclear': 'nuclear', '5_island': 'island', '3_,': ',', '4_the': 'the', '11_set': 'set', '6_of': 'of', '7_stability': 'stability', '8_is': 'is', '9_a': 'a', '10_predicted': 'predicted', '12_of': 'of', '13_isotopes': 'isotopes', '15_of': 'of', '19_elements': 'elements', '16_superheavy': 'superheavy', '20_that': 'that', '22_have': 'have', '21_may': 'may', '23_considerably': 'considerably', '24_longer': 'longer', '25_half': 'half', '28_than': 'than', '30_isotopes': 'isotopes', '29_known': 'known', '32_of': 'of', '34_elements': 'elements', '33_these': 'these', '35_.': '.'}
Node POS tags:
{'0_in': 'IN', '2_physics': 'NN', '1_nuclear': 'JJ', '5_island': 'NNP', '3_,': ',', '4_the': 'DT', '11_set': 'NN', '6_of': 'IN', '7_stability': 'NN', '8_is': 'VBZ', '9_a': 'DT', '10_predicted': 'VBN', '12_of': 'IN', '13_isotopes': 'NNS', '15_of': 'IN', '19_elements': 'NNS', '16_superheavy': 'JJ', '20_that': 'WDT', '22_have': 'VB', '21_may': 'MD', '23_considerably': 'RB', '24_longer': 'JJR', '25_half': 'NNS', '28_than': 'IN', '30_isotopes': 'NNS', '29_known': 'VBN', '32_of': 'IN', '34_elements': 'NNS', '33_these': 'DT', '35_.': '.'}
edge labels:
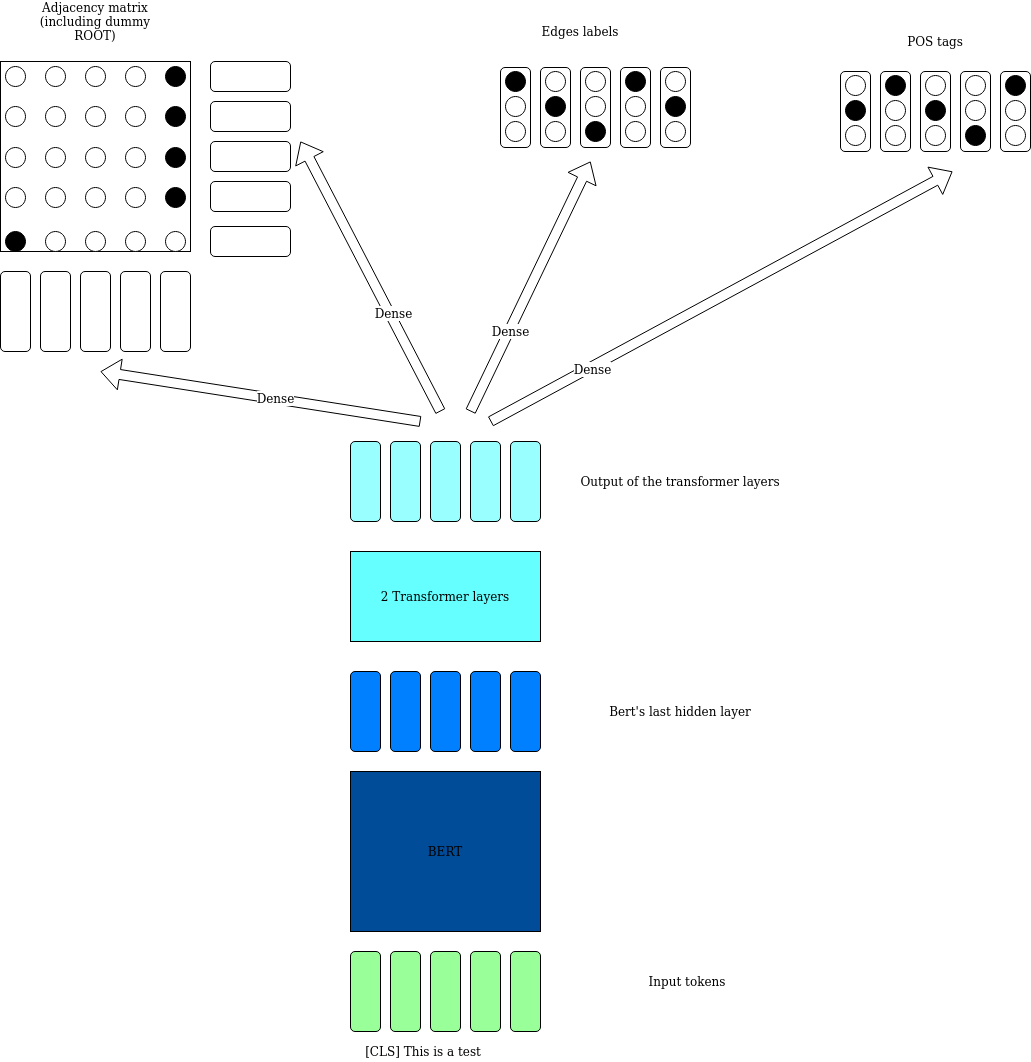
{('0_in', '2_physics'): 'case', ('2_physics', '5_island'): 'nmod', ('1_nuclear', '2_physics'): 'amod', ('5_island', '11_set'): 'nsubj', ('3_,', '2_physics'): 'punct', ('4_the', '5_island'): 'det', ('6_of', '7_stability'): 'case', ('7_stability', '5_island'): 'nmod', ('8_is', '11_set'): 'cop', ('9_a', '11_set'): 'det', ('10_predicted', '11_set'): 'amod', ('12_of', '13_isotopes'): 'case', ('13_isotopes', '11_set'): 'nmod', ('15_of', '19_elements'): 'case', ('19_elements', '13_isotopes'): 'nmod', ('16_superheavy', '19_elements'): 'amod', ('20_that', '22_have'): 'nsubj', ('22_have', '19_elements'): 'acl:relcl', ('21_may', '22_have'): 'aux', ('23_considerably', '24_longer'): 'advmod', ('24_longer', '25_half'): 'amod', ('25_half', '22_have'): 'obj', ('28_than', '30_isotopes'): 'case', ('30_isotopes', '22_have'): 'nmod', ('29_known', '30_isotopes'): 'amod', ('32_of', '34_elements'): 'case', ('34_elements', '30_isotopes'): 'nmod', ('33_these', '34_elements'): 'det', ('35_.', '11_set'): 'punct'}The stylized architecture is depicted below
While the full code is found in the file model.py
The system is trained with an Adam optimizer using 5e-5 as step size.
The two transformer layers seem to add stability to training: without the the system ends up stuck in a local minimum after a few epochs.
The system is trained on the UG dataset en_gum-ud
The results of the system are quite good. A comparison with a non BERT-based method can be found in the excellent paper Nguyen and Vespoor: Bert seems to add 6% points in UAS and 7 in LAS. Curiously the POS score is increased by only 1 percentage point.
| Set | UAS | LAS | POS |
|---|---|---|---|
| Dev | 0.91 | 0.88 | 0.96 |
| Test | 0.9 | 0.87 | 0.95 |
A pretrained BERT language model seems to be improving upon LSTM based training. By quite a lot!