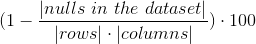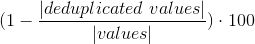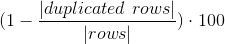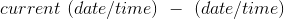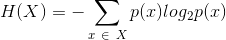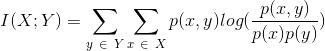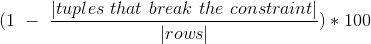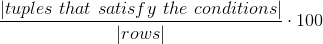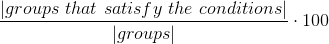A data quality tool (docs).
Haychecker provides both a script to be called by submitting it to spark-submit or to be run with the python3 interpreter, and a library. The script can be found in the root directory, named hay_checker.py, the library can be installed with pip either online or by running setup tools and creating a package installable with pip locally (the setup.py file can be found in the root directory). By submitting the script to spark-submit, spark will be used to import the data and perform computations, while by using the python interpreter spark will be used to import the data, and pandas to perform computations.
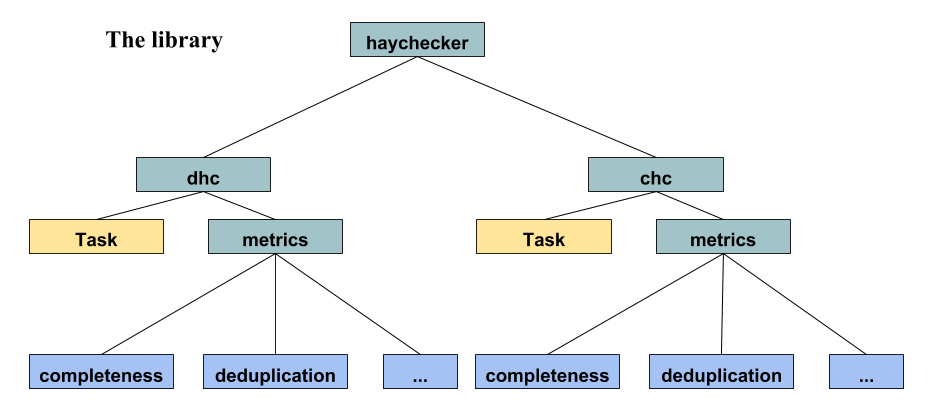
The library has 2 main packages, chc and dhc, which respectively stand for centralized hay checker and distributed hay checker, they are basically two "symmetric" libraries themselves, with the same exact API. Those libraries are based on a single Task class, which is a container for metrics that will/can be computed later or on different dataframes, and a number of functions (metrics) which will either compute and return results when called or return a Task, to be later run or expanded by adding more metrics/tasks to it.
The point of the Task class is to 1) allow defining things once, but to use those defined metrics multiple times, 2) trying to optimize/combine metrics that can be run together in a single call, instead of calling them one at a time, when possible.
- haychecker, (directory with source)
- run_tests.sh (to run all tests, which are in haychecker/test)
- hay_checker.py (the script that can be either submitted to spark or run in python3)
- docs (documentation)
- examples (examples with source)
- setup.py (setup script for setuptools)
- dist, which contain a source archive and a built distribution (.whl), installable with "pip3 install .tar.gz" or "pip3 install .whl"
- other stuff (license, figures, readme)
(check the setup.py file for what version of the requirements is going to be installed, we set up kinda recent versions to be required of pyspark, pandas, etc.)
- pip3 install haychecker, to download from PyPI or
- pip3 install dist/haychecker-<version>.tar.gz, or
- pip3 install dist/haychecker-<version>-py3-none-any.whl
Docs can be found at this project github page, the same docs can be found in the directory "docs" of the project. Examples also has some examples with source code, but the README should suffice to give a general know-how.
These instructions will provide a simple guide on how to use the tool and the prerequisites needed for both distributed and centralized versions of hay checker
Requirements:
hadoop
spark
pyspark
pandas
sklearn
The script hay_checker.py can be used by calling
spark-submit your-spark-parameters hay_checker.py --config <path to config.json>or
python3 hay_checker.py --config <path to config.json>to use the distributed or centralized versions, respectively. The distributed version will use spark for data importing and computation, the centralized version will use spark for data importing and pandas for computation.
Input
A .json configuration file, where the following values can be provided:
| table | Path to the data, see below this table for more details |
| inferSchema | boolean, spark inferSchema argument for csv import, setting this to true will also allow casting of columns which are strings when confronted with values having a numeric type, if false such casting is not performed, and instead an error is raised |
| delimiter | string, spark delimiter argument for csv import, optional, default="," |
| header | boolean, spark inferSchema argument for csv import, optional, default=true |
| output | a string with path to output file |
| verbose | boolean, if true configuration options are printed to the screen, optional, default=false |
| metrics | a list of metric objects, composed by the following fields: - "metric": a string specifying the name of the metric - metric arguments, in key:value format |
Possible ways to provide data with the "table" parameter in the config json:
- local file, i.e. file:///home/myname/mydata.csv
- file in the hdfs, such as /mydata.csv
- a table through jdbc, the string must specify the jdbc url first and the table name second, separated by space; i.e. "jdbc://path cooltable". Note that the url must always begin with "jdbc"
- data from the web, currently only the json and csv format are supported, the url must always begin with "http", and it must contain either csv or json respectively
- a table from the hdfs, the data importer will assume that a table has been passed if: the "table" string does not begin with "http" or "jdbc", and it does not terminate with "parquet", "json", "csv".
Local and hdfs file import currently support parquet, csv and json files; the file must terminate with ".parquet", ".csv" or ".json".
config-example.json in the examples/resources directory is provided as an example.
Output
A .json file containing scores of input metrics
Simple examples of metrics usage. More examples and source code can be found in examples folder. Metrics functions all have allow_casting enabled (see the inferSchema in the above table if you are confused by this).
Centralized version
Import a dataframe
import pandas as pd
df = pd.read_csv("examples/resources/employees.csv")
pd.set_option('max_columns', 10)
print(df)Output
lastName firstName title birthDate hireDate \
0 Davolio Nancy Sales Representative 1948/12/08 1992/05/01
1 Fuller Andrew Vice President, Sales 1952/02/19 1992/08/14
2 Leverling Janet Sales Representative 1963/08/30 1992/04/01
3 Peacock Margaret Sales Representative 1937/09/19 1993/05/03
4 Buchanan Steven Sales Manager 1955/03/04 1993/10/17
5 Suyama Michael Sales Representative 1963/07/02 1993/10/17
6 King Robert Sales Representative 1960/05/29 1994/01/02
city region reportsTo salary
0 Seattle WA 2.0 2000
1 Tacoma WA NaN 3000
2 Kirkland WA 2.0 2000
3 Redmond WA 2.0 2000
4 London NaN 2.0 2500
5 London NaN 5.0 2000
6 London NaN 5.0 2000
Distributed version
Create a spark session
from pyspark.sql import SparkSession
spark = SparkSession.builder.appName("Deduplication_approximated_example").getOrCreate()Import a dataframe
df = spark.read.format("csv").option("header", "true").load("examples/resources/employees.csv")
df.show()Output
+---------+---------+--------------------+----------+----------+--------+------+---------+------+
| lastName|firstName| title| birthDate| hireDate| city|region|reportsTo|salary|
+---------+---------+--------------------+----------+----------+--------+------+---------+------+
| Davolio| Nancy|Sales Representative|1948/12/08|1992/05/01| Seattle| WA| 2| 2000|
| Fuller| Andrew|Vice President, S...|1952/02/19|1992/08/14| Tacoma| WA| null| 3000|
|Leverling| Janet|Sales Representative|1963/08/30|1992/04/01|Kirkland| WA| 2| 2000|
| Peacock| Margaret|Sales Representative|1937/09/19|1993/05/03| Redmond| WA| 2| 2000|
| Buchanan| Steven| Sales Manager|1955/03/04|1993/10/17| London| null| 2| 2500|
| Suyama| Michael|Sales Representative|1963/07/02|1993/10/17| London| null| 5| 2000|
| King| Robert|Sales Representative|1960/05/29|1994/01/02| London| null| 5| 2000|
+---------+---------+--------------------+----------+----------+--------+------+---------+------+
In the following examples all imports are of the form from haychecker.dhc.metrics import <metric> For the centralized version all metric calls are identical, while import has the form from haychecker.chc.metrics import <metric>
Measures how complete is the dataset by counting which entities are not missing (Nan or Null/None) in a column or in the whole table.
- column:
- table:
method metrics.completeness(columns=None, df=None)
Arguments
| columns | Columns on which to run the metric, None to run the completeness metric on the whole table. |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later. |
Example
from haychecker.dhc.metrics import completeness
r1, r2 = completeness(["region", "reportsTo"], df)
print("Completeness region: {}, completeness reportsTo: {}".format(r1, r2))Output
Completeness region: 57.14285714285714, completeness reportsTo: 85.71428571428571
Measures how many values are duplicated within a column/dataset, NaN and Null values are considered duplicated.
- column:
- table:
method metrics.deduplication(columns=None, df=None)
Arguments
| columns | Columns on which to run the metric, None to run the deduplication metric on the whole table. |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later |
Example
from haychecker.dhc.metrics import deduplication
r1, r2 = deduplication(["title", "city"], df)
print("Deduplication title: {}, deduplication city: {}".format(r1, r2))Output
Deduplication title: 42.857142857142854, deduplication city: 71.42857142857143
Deduplication metric implementation using approximated count of distinct values, faster then the standard deduplication implementation.
method metrics.deduplication_approximated(columns=None, df=None)
Arguments
| columns | Columns on which to run the metric, differently from deduplication, this cannot be run "on the whole table", checking deduplication at a row level, but only on 1 column at a time, meaning that the columns parameter cannot be None, and must always contain at least one column. |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containingthis metric to be run later |
Example
from haychecker.dhc.metrics import deduplication_approximated
r1, r2 = deduplication_approximated(["title", "city"], df)
print("Deduplication_approximated title: {}, deduplication_approximated city: {}".format(r1, r2))Output
Deduplication_approximated title: 42.857142857142854, deduplication_approximated city: 71.42857142857143
Reflects how up-to-date the dataset is with respect to the given date/time.
- column
! In the centralized version of the library the strftime directives are used to express the date/time format arguments, while in the distributed version the simple date format directives are used.
method metrics.timeliness(columns, value, df=None, dateFormat=None, timeFormat=None)
Arguments
| columns | Columns on which to run the metric, columns of type string will be casted to timestamp using the dateFormat or timeFormat argument |
| value | Value used to run the metric, confronting values in the specified columns against it |
| dateFormat | Format in which the value (and values in columns, if they are of string type) are; used to cast columns if they contain dates as strings. Either dateFormat or timeFormat must be passed, but not both |
| timeFormat | Format in which the value (and values in columns, if they are of string type) are; used to cast columns if they contain dates as strings. Either dateFormat or timeFormat must be passed, but not both |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later |
If a dateFormat is passed, only days are considered for the comparison, hours, minutes, seconds, etc. are ignored.
If a timeFormat is passed and it contains tokens that represent years, months, days, or anything related to dates, the comparison is made considering "everything". On the contrary, the comparison will only consider hours, minutes and seconds, allowing for a comparison between a value which is just a time, like 23:54:10 and timestamps which contain also years, months, etc; if we want to only compare by times while also having years/months/days in our timestamp.
Example
from haychecker.dhc.metrics import timeliness
r1 = timeliness(["birthDate"], dateFormat="yyyy/MM/dd", df=df, value="1960/10/22")[0]
print("Timeliness birthDate with respect to date 1960/10/22: {}".format(r1))Output
Timeliness birthDate with respect to date 1960/10/22: 71.42857142857143
Measures how fresh is the dataset by taking the average distance of the tuples from the current date/time. Results are returned either in days or seconds, depending on what format is passed, dateFormat for days and timeFormat for seconds.
- column
! In the centralized version of the library the strftime directives are used to express the date/time format arguments, while in the distributed version the simple date format directives are used.
method metrics.freshness(columns, df=None, dateFormat=None, timeFormat=None)
Arguments
| columns | Columns on which to run the metric, columns of type string will be casted to timestamp using the dateFormat or timeFormat argument. |
| dateFormat | Format in which the values in columns are if those columns are of type string; otherwise they must be of type date or timestamp. Use this parameter if you are interested in a result in terms of days. Either dateFormat or timeFormat must be passed, but not both. |
| timeFormat | Format in which the values in columns are if those columns are of type string; otherwise they must be of type timestamp. Use this parameter if you are interested in results in terms of seconds. Either dateFormat or timeFormat must be passed, but not both. |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later |
If a dateFormat is passed, the average distance in days is returned, without considering hours, minutes, seconds, etc.
If a timeFormat is passed and it contains tokens that represent years, months, days, or anything related to dates, the average distance in seconds is returned, also considering years, months and days. If the timeFormat does not contain anything related to dates, the average distance will be computed without considering years, months and days. This allows for computing the average distance only considering the hours, minutes, and seconds in our timestamps, if we are interested in how distant they are from "now" as a time and not as a date.
Example
from haychecker.dhc.metrics import freshness
r1 = freshness(["birthDate"], dateFormat="yyyy/MM/dd", df=df)[0]
print("Freshness birthDate: {}".format(r1))Output
Freshness birthDate: 23434.571428571428 days
Shannon entropy of a column.
- column X
method metrics.entropy(column, df=None)
Arguments
| column | Columns on which to run the metric |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containin gthis metric to be run later |
Null and NaN are ignored for computing results.
Example
from haychecker.dhc.metrics import entropy
r1 = entropy("firstName", df)[0]
print("Entropy firstName: {}".format(r1))Output
Entropy firstName: 2.807354922057604
- two columns X and Y
method metrics.mutual_info(when, then, df=None)
Arguments
| when | First column on which to compute MI |
| then | Second column on which to compute MI |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containin gthis metric to be run later |
Null and NaN are ignored for computing results (pairs containing Nans or Nulls are discarded).
Example
from haychecker.dhc.metrics import mutual_info
r1 = mutual_info("title", "salary", df)[0]
print("Mutual information title/salary: {}".format(r1))Output
Mutual information title/salary: 0.7963116401738128
After filtering data on conditions, measures how many tuples satisfy/break a given functional dependency, a functional dependency is a dependency where values of one set of columns imply values of another set of columns.
- table
method metrics.constraint(when, then, conditions=None, df=None)
Arguments
| when | A list of columns in the df to use as the precondition of a functional constraint. No column should be in both when and then |
| then | A list of columns in the df to use as the postcondition of a functional constraint. No column should be in both when and then |
| conditions | Conditions on which to filter data before applying the metric |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later |
If any condition would lead to a comparison between a column of string type and a number, if casting is allowed (allow_casting in the Task class constructor, or inferSchema in the config.json file), there will be an attempt to cast the column to a numeric type.
Conditions are expressed as dictionaries, are defined on one column at a time, and allow for the following operators: "gt", "lt" and "eq"; value should be a string or a number. Conditions in the list we pass as an argument are intended to be joined by AND.
condition1 = {"column": "salary", "operator": "gt", "value": 2100}Example
from haychecker.dhc.metrics import constraint
r1 = constraint(["title"], ["salary"], df=df)[0]
print("Constraint title/salary: {}".format(r1))Output
Constraint title/salary: 100.0
Measures how many values satisfy a rule
- table
method metrics.rule(conditions, df=None)
Arguments
| conditions | Conditions on which to run the metric |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later |
If any condition would lead to a comparison between a column of string type and a number, if casting is allowed (allow_casting in the Task class constructor, or inferSchema in the config.json file), there will be an attempt to cast the column to a numeric type.
Conditions are expressed as dictionaries, are defined on one column at a time, and allow for the following operators: "gt", "lt" and "eq"; value should be a string or a number. Conditions in the list we pass as an argument are intended to be joined by AND.
condition1 = {"column": "salary", "operator": "gt", "value": 2100}Example
from haychecker.dhc.metrics import rule
condition1 = {"column": "salary", "operator": "gt", "value": 2100}
conditions = [condition1]
r1 = rule(conditions, df)[0]
print("Rule salary>2100: {}".format(r1))Output
Rule salary>2100: 28.57142857142857
Measures how many values satisfy a rule by filtering data on conditions, and then checking the percentage of groups passing the having conditions, which are conditions on aggregations on the groups.
- table
method metrics.contains_date(format)
Arguments
| columns | Columns on which to run the metric, grouping data |
| conditions | Conditions on which to run the metric, filtering data before grouping, can be None |
| having | Conditions to apply to groups |
| df | Dataframe on which to run the metric, None to have this function return a Task instance containing this metric to be run later |
If any condition would lead to a comparison between a column of string type and a number, if casting is allowed (allow_casting in the Task class constructor, or inferSchema in the config.json file), there will be an attempt to cast the column to a numeric type.
Conditions are expressed as dictionaries, are defined on one column at a time, and allow for the following operators: "gt", "lt" and "eq"; value should be a string or a number. Conditions in the list we pass as an argument are intended to be joined by AND.
condition1 = {"column": "salary", "operator": "gt", "value": 2100}Havings are conditions expressed on aggregations of certain columns, as conditions, they are defined as dictionaries, specified on one column at a time and intended as joined by an AND operator. The operators "gt", "lt" and "eq" are allowed, and the aggregators "count", "min", "max", "avg", "sum" can be used. To perform a "count(*)", simply provide "count" as an aggregator, and "*" as column.
having1 = {"column": "*", "operator": "gt", "value": 1, "aggregator": "count"}Example
from haychecker.dhc.metrics import grouprule
condition1 = {"column": "city", "operator": "eq", "value": "London"}
conditions = [condition1]
having1 = {"column": "*", "operator": "gt", "value": 1, "aggregator": "count"}
havings = [having1]
r1 = grouprule(["title"], havings, conditions, df)[0]
print("Grouprule groupby \'title\' where city=\'London\' having count * > 1: {}".format(r1))Output
Grouprule groupby 'title' where city='London' having count * > 1: 50.0
The Task class acts as a container for metrics, allowing to store them and add more of them programmatically, a Task can be either instantiated empty (Task()) or by passing a list of metrics, where each metric is a dict defining parameters as in the config.json file. Tasks can be added to other tasks, (basically appending the metrics contained in one to the other); moreover, a metric function called without a dataframe will return a Task containing the metric itself, effectively allowing to add more metrics to a Task instance in a natural way. Metrics that can be run in the same call/aggregation/pass on data will be run in such a way, otherwise they will be run internally one at a time.
class task.Task(metrics_params=[], allow_casting=True)
Arguments
| metrics_params | List of metrics, each metric is a dictionary mapping params of the metric to a value, as in the json config file |
| allow_casting | If a column not of the type of what it is evaluated against (i.e. a condition checking for column 'x' being gt 3.0, with the type of 'x' being string) should be casted to the type of the value it is checked against. If casting is not allowed the previous example would provoke an error, (through an assertion); default is True |
Example
If in a metric function call a dataframe df is not specified, a Task instance with input parameters is returned.
from haychecker.dhc.task import Task
from haychecker.dhc.metrics import *
# create an empty Task
task = Task()
# the metric function returns a Task instance because it has not been called with a df
task1 = completeness(["region", "reportsTo"])
task2 = completeness(["city"])
task.add(task1)
task.add(task2)
# we can also add tasks is a more natural and concise way
# the add operator also allows chaining
task.add(deduplication(["title", "city"]))
task.add(deduplication(["lastName"]))
# run all metrics contained in the task
# the method will return a list of dictionaries which are the metrics
# contained in the Task instance, where each metric has also a "scores" field added, containing
# related scores
result = task.run(df)
r1, r2 = result[0]["scores"]
r3 = result[1]["scores"][0]
print("Completeness region: {}, completeness reportsTo: {}, completeness city: {}".format(r1, r2, r3))
r1, r2 = result[2]["scores"]
r3 = result[3]["scores"][0]
print("Deduplication title: {}, deduplication city: {}, deduplication lastName: {}".format(r1, r2, r3))Output
Completeness region: 57.14285714285714, completeness reportsTo: 85.71428571428571, completeness city: 100.0
Deduplication title: 42.857142857142854, deduplication city: 71.42857142857143, deduplication lastName: 100.0
- Jacopo Gobbi
- Kateryna Konotopska
This project is licensed under the MIT License - see the LICENSE file for details