An R Package for the analysis of RNA editing data from studies utilizing whole genome sequencing and RNA-seq from the same individual(s).
editTools was made out of the necessity to create a reproducible means to analyze RNA editing data. Candidate RNA editing detection is most often performed using scripts to analyze variant call format (VCF) data, however such scripts are rarely published leading to the possibility that variation in RNA editing results may be partly due to subtle differences in script design. This R package provides the means to analyze VCF data using standard methodology inspired from previous studies, and do so within the R framework where powerful graphical tools can be utilized to best explore the data.
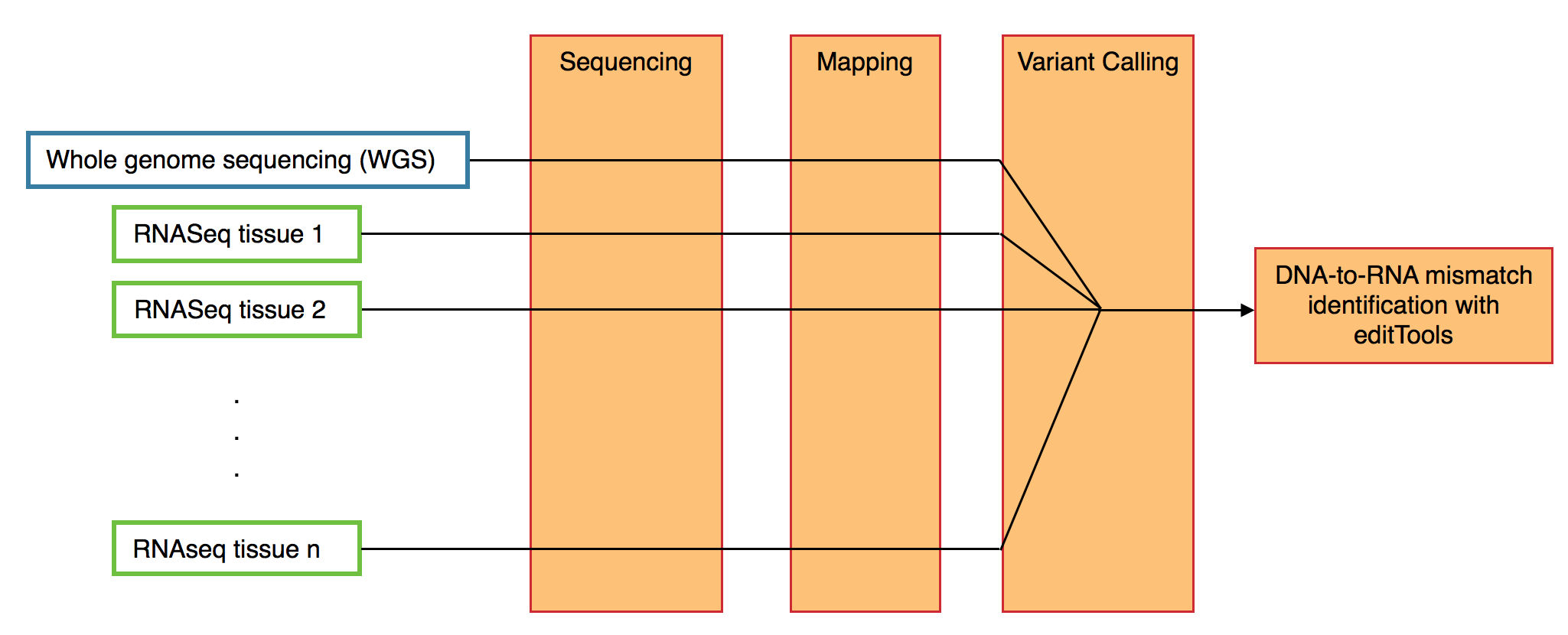
From a single individual, editTools can analyze Whole genome sequencing (WGS) as well as RNAseq from any number of tissues. Input for editTools can be prepared a number of ways (allowing for differential sequencing, mapping, and variant calling techniques).
An example of how I prepare input for editTools can be found here
editTools contains compiled code and relies on the Rcpp package and C++11. Please note that editTools has only been tested on Mac and Linux machines and it would not be surprising if it's missing components or installation instructions needed for Windows.
If using GNU version 4.7 or later, specify that you want to install editTools using C++11 with:
Sys.setenv("PKG_CXXFLAGS" = "-std=c++11")Otherwise if using an older version of the GNU, use:
Sys.setenv("PKG_CXXFLAGS" = "-std=c++0x")Failure to use one of the above commands before attempting to install editTools will result in fatal compilation errors.
If the devtools package is not already installed, then install it with install.packages("devtools"), then install the editTools package with:
devtools::install_github("funkhou9/editTools")The only current requirement for editTools is that the input (resulting from the Variant Calling software of choice), must be in VCF format and contain the following header information.
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT <DNA> <RNA1> <RNA2> <RNA3> ...
where <DNA> corresponds to your WGS sample, and <RNA1>, <RNA2>, <RNA3> ... correspond to any number of RNA samples. Currently, the FORMAT field must contain DP (total depth),DV (variant depth), and SP (strand-bias p-value). Steps for establishing those fields can be found here
VCF format is (currently, to my knowledge) unaware of strand specificity. In other words, all bases reported in a VCF file correspond to the TOP strand of the genome. This presents a challenge when calling transcriptome variants. For RNA editing discovery, this means that one cannot distinguish an A-to-G (DNA-to-RNA) mismatch from a T-to-C mismatch, since a single cDNA read will contain both the G allele and the C allele, and knowledge of which strand that cDNA read was generated from is absent from VCF files. To workaround this and have the ability to distinguish (for example) A-to-G mismatches from T-to-C mismatches, the user can prepare two VCF files, one containing variants on plus-strand transcripts for each <RNA> sample, and one containing variants on minus-strand transcripts for each <RNA> sample. In each case, the same <DNA> sample is provided.
If two VCF files are prepared (see above), then identification of DNA-to-RNA mismatches can be performed with:
edits <- editTools::find_edits(<plus.vcf>, <minus.vcf>, names = c("Brain", "Liver", "Heart"))<plus.vcf>VCF file containing plus-strand transcripts for each RNA sample (a character string).<minus.vcf>VCF file containing minus-strand transcripts for each RNA sample (a character string).namesDesired names for RNA samples in the order that they appear in the VCF file (a character vector).
Without supplying additional arguments, by default editTools will scan both VCF files and identify candidate RNA edited loci according to criteria largely inspired by Chen et al. 2014, in which a candidate RNA edited site must have:
- A genotype that is homozygous according to 95% of the gDNA reads
- A genotype sequencing depth of at least 10 gDNA reads
- An "editing depth" (the cDNA sequencing depth in support of an alternative base to what is indicated by gDNA) of at least 5 cDNA reads
Additionally, editTools will ignore all sites containing indels in either the gDNA or any of the cDNA samples as a means to only search for single nucleotide mismatches, and will require that the gDNA sample and the cDNA sample supporting a mismatch both have a phred-scaled strand bias p-value of at least 20.
For information on all advanced options to tweak mismatch idenfication parameters, see:
?editTools::find_editsedits is an object of class "edit_table", and initially contains two fields: AllSites and Tissues.
-
Allsitesis a data.frame with candidate RNA editing events in the rows with the following columns:IDA numerical ID associated with the tissue specific candidate editing event.ChrA character representing the chromosome of the edit.PosA numeric representing the chromosomal position of the edit.StrandA character representing the strand of the edited transcript ("+" or "-")MismatchA character representing the mismatch (eg. "AtoG")DNA_depthA numeric representing the total DNA sample sequencing depthDNA_variant_depthA numeric representing the DNA sample sequencing depth that is in support of a variantRNA_depthA numeric representing the total RNA sample sequencing depthRNA_mismatch_depthA numeric representing the RNA sample sequencing depth in support of a DNA-to-RNA mismatchRNA_edit_fracA numeric. Simply theRNA_mismatch_depth/RNA_depthPhred_strand_biasA numeric representing the phred-scaled probabilities of strand-bias.Ave_MQA numeric representing the average mapping quality at this site across all samples.TissueA character indicating which tissue (named withnamesargument) the event was found in.
-
Tissuesis a list with the number of elements equal to the number of tissues studied. Each element is a data.frame with mismatch types (A-to-G) in rows and the following columns:MismatchA character with the type of mismatchFreqThe number of events found of the specified type of mismatchPropThe proportion of mismatches belonging to the specified type
Without importing data from RepeatMasker or Variant Effect Predictor, the edit_table object will contain no repetitive element or gene annotation, respectively. editTools facilitates the merging of edit_table objects with Repeatmasker genomic datasets (example) or Variant effect predictor output.
editTools implements a pseudo binary search approach to find the positions of each DNA-to-RNA mismatch within a repeatmasker dataset. In theory, this should allow for very large edit_table objects to be searched among large genomic repeatmasker datasets.
To add RepeatMasker annotation to the edit_table object edits, simply use:
edits <- editTools::add_repeatmask(edits, <rm.out>)where <rm.out> is RepeatMasker output from a genomic dataset, such as the one provided in the example above.
For adding gene annotation, likewise use:
edits <- editTools::add_VEP(edits, <vep.out>)where <vep.out> is VEP output generated from the positions of each mismatch.
To generate
<vep.out>, input for VEP can be generated withwrite_vep(edits)
Currently VEP outputs containing the fields
Consequence,BIOTYPE,STRAND,SYMBOL,Gene,TREMBL,IMPACT,Amino_acids, andSIFTare all required, although editTools will soon support VEP outputs containing fewer/different fields.
A number of plotting tools are included, such as an S3 method for plot() that can be used to generate a "DNA-to-RNA mismatch count histogram".
plot(edits)will generate a histogram showing all DNA-to-RNA mismatch types and their counts for all tissue samples studied.
plot(edits, field = "RepSites")will only construct a histogram considering mismatches within repetitive elements.
Information for more plotting techniques can be found by using:
edit_prop_plot(edits)will produce a plot showing the distribution of editing levels among the dataset.
- Monumental changes to program design - Incorporate new input types. The ability to process BAM files instead of VCF files could greatly enhance editTools ease of use. For instance, separating plus-strand alignments from minus-strand alignments would no longer be needed by the user. But how would the identification of gDNA and cDNA variants be affected?