Deep learning has been playing an important role in modern agriculture, especially in plant pathology using leaf images where convolutional neural networks (CNN) are attracting a lot of attention. In this paper, we start our study by surveying current deep learning approaches for plant identification and disease classification. We categorise the approaches into multi-model, multi-label, multi-output, and multi-task, in which different backbone CNNs can be employed. Furthermore, based on the survey of existing approaches in plant pathology and the study of available approaches in machine learning, we propose a new model named Generalised Stacking Multi-output CNN (GSMo-CNN). To investigate the effectiveness of different backbone CNNs and learning approaches, we conduct an intensive experiment on three benchmark datasets Plant Village, Plant Leaves, and PlantDoc. The experiment results demonstrate that InceptionV3 can be a good choice for a backbone CNN as its performance is better than AlexNet, VGG16, ResNet101, EfficientNet, MobileNet, and a custom CNN developed by us. Interestingly, there is empirical evidence to support the hypothesis that using a single model for both tasks can be comparable or better than using two models, one for each task. Finally, we show that the proposed GSMo-CNN achieves state-of-the-art performance on three benchmark datasets.
If you find our work helpful, please kindly cite our paper as:
@article{10.1145/3639816,
author = {Yao, Jianping and Tran, Son N. and Garg, Saurabh and Sawyer, Samantha},
title = {Deep Learning for Plant Identification and Disease Classification from Leaf Images: Multi-Prediction Approaches},
year = {2024},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
issn = {0360-0300},
url = {https://doi.org/10.1145/3639816},
doi = {10.1145/3639816},
note = {Just Accepted},
journal = {ACM Comput. Surv.},
month = {jan},
keywords = {plant pathology, convolutional neural networks, multi-prediction, deep learning, plant identification, leaf disease classification}
}
JIANPING YAO & SON N. TRAN*
- contact: sn.tran@utas.edu.au/sontn.fz@gmail.com
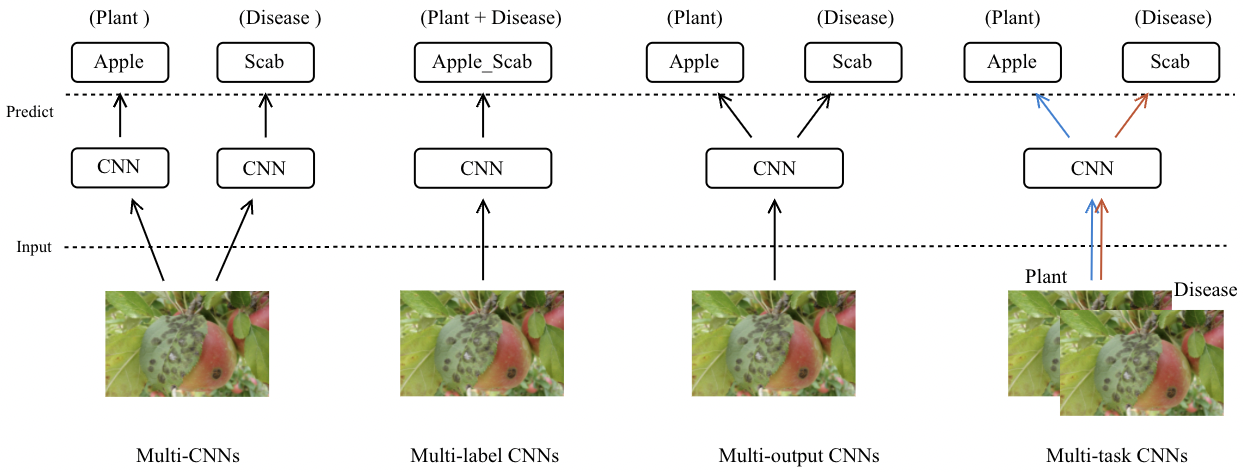
CNNs approaches for plant identification and Disease classification:
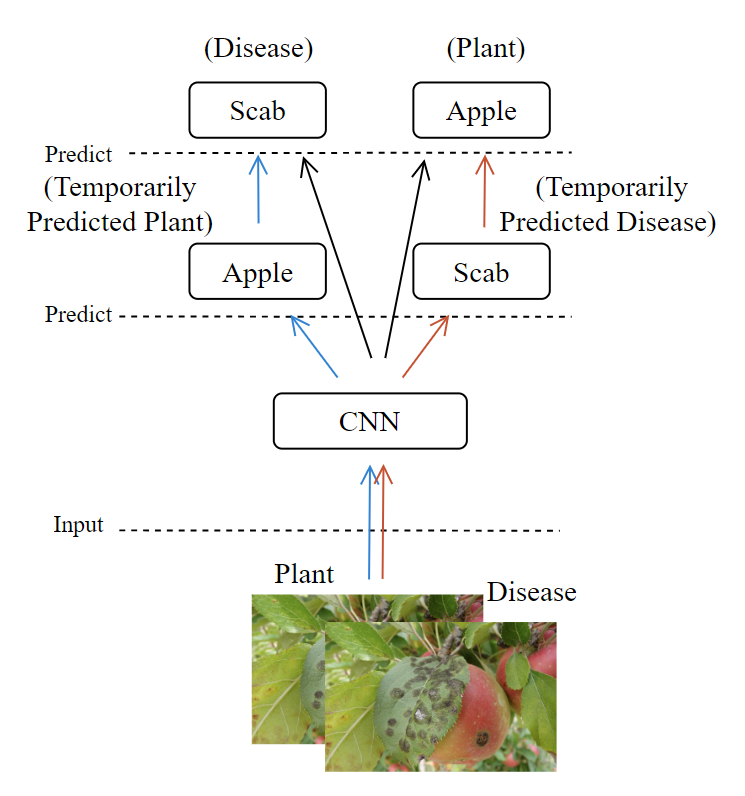
Our New Model Structure:
- Python >= 3.7
- h5py = 3.1.0
- Matplotlib = 3.5.1
- Numpy = 1.19.2
- opencv-contrib-python = 4.5.5.64
- opencv-python = 4.5.5.64
- Pandas = 1.4.2
- scikit-learn = 1.0.2
- tensorflow-gpu = 2.4.1 or 2.6.0
- Imutils = 0.5.4
- PyTorch = 1.11.0
- torchvision = 0.12.0
Please modify the parameters in main.py, then run python main.py.
Please download the related datasets and put them in the path of dataset_dir (e.g., /home/user/Leaf_diseases/Datasets/).
-
Plant Village: i.e.,
plant_village(https://data.mendeley.com/datasets/tywbtsjrjv/1). Please downloadPlant_leaf_diseases_dataset_without_augmentation.zip(828MB), then unzip it and rename the folder name toplant_village(Note: please remove sub-folderBackground_without_leavesinPlant_leave_diseases_dataset_without_augmentation). Ensure the path looks like this:/home/user/Leaf_diseases/Datasets/plant_village/Plant_leave_diseases_dataset_without_augmentation/Apple___Apple_scab. -
Plant Leaves: i.e.,
plant_leaves(https://data.mendeley.com/datasets/hb74ynkjcn/1). Please clickdownload All(6718 MB) and put them in a folder which should be namedplant_leaves. Combine the sub-folder name and sub-sub-folder names. E.g.,Alstonia Scholaris (P2)hasdiseased&healthy, then rename them toAlstoniaScholaris_diseased&AlstoniaScholaris_healthy. Ensure the path looks like this:/home/user/Leaf_diseases/Datasets/plant_leaves/AlstoniaScholaris_diseased. -
PlantDoc-0.2: i.e.,
PlantDoc(https://github.com/pratikkayal/PlantDoc-Dataset) Please downloadPlantDoc-Datasetand rename the folder name toPlantDoc_Dataset, Please remove the folderTomato two spotted spider mites leafwhich has only two pictures in thetestfolder first. Please copy all items from thetestfolder to thetrainfolder (Note: the image names are case sensitive, and may not be suitable for some Windows environments). Ensure the path looks like this:/home/user/Leaf_diseases/Datasets/PlantDoc_Dataset/train. -
PlantDoc-1.0: i.e.,
PlantDoc_original(https://github.com/pratikkayal/PlantDoc-Dataset) Please downloadPlantDoc-Datasetand rename the folder name toPlantDoc_original, Please remove the folderTomato two spotted spider mites leafwhich has only two pictures in thetestfolder first. Please keep thetestfolder and thetrainfolder (Note: the image names are case sensitive, and may not be suitable for some Windows environments). Ensure the path looks like this:/home/user/Leaf_diseases/Datasets/PlantDoc_original/trainand/home/user/Leaf_diseases/Datasets/PlantDoc_original/test.
| Parameter | Description |
|---|---|
dataset_dir |
The path of the dataset, e.g., /home/user/Leaf_diseases/Datasets/. |
save_path |
The path of results and model will be saved, e.g., /home/user/Leaf_diseases/test_model/ |
item |
Dataset name, Options: plant_village, plant_leaves, PlantDoc & PlantDoc_original. |
obj |
Approach name, Options: multi_model, multi_output, new_model, multi_label & cross_stitch. |
model_name |
backbone name, Options: CNN, AlexNet, VGG, ResNet, EfficientNet, Inception & MobileNet. |
saveornot |
Save or Not save the results , Options: save & not. |
fig_size |
Fig size of the images, default=256. |
INIT_LR |
Learning rate, default = 0.001 |
op_z |
Optimizer, default = Adamax. |
TF_weights |
Transfer Learning weights, Options: None & imagenet |
bat_si |
Batch size, default = 16 |
epo |
Epoch, default = 10000 |
times |
The times’ amount of the model will run, default = 10 |
p_t_w |
The p_t weight of our new method, default = 0.1 |
d_t_w |
The d_t weight of our new method, default = 0.1 |
p_w |
The p weight of our new method, default = 0.4 |
d_w |
The d weight of our new method, default = 0.5 |
balance_weight |
A list of the balance weight in our method, i.e., [p_t_w, d_t_w, p_w, d_w] |
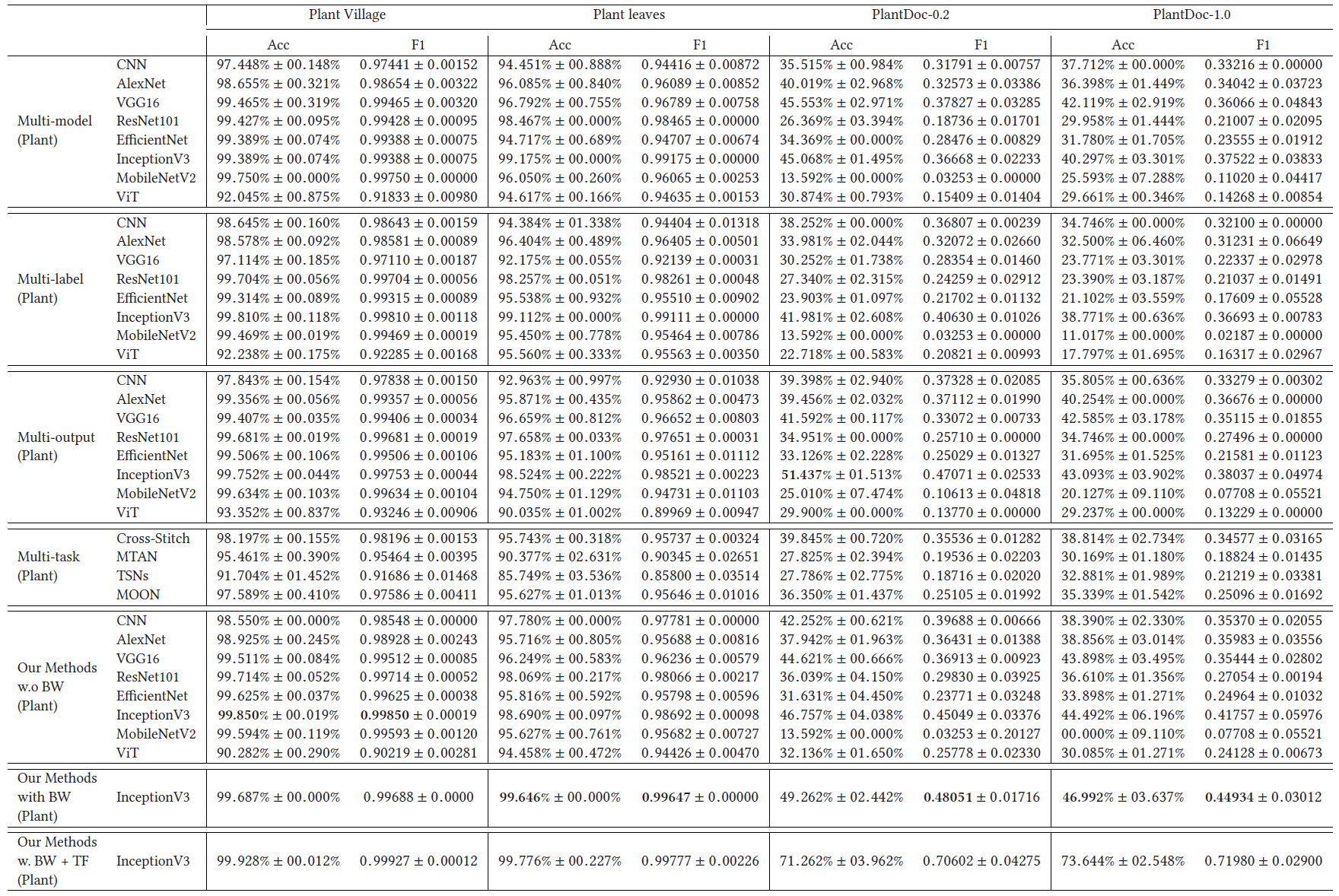
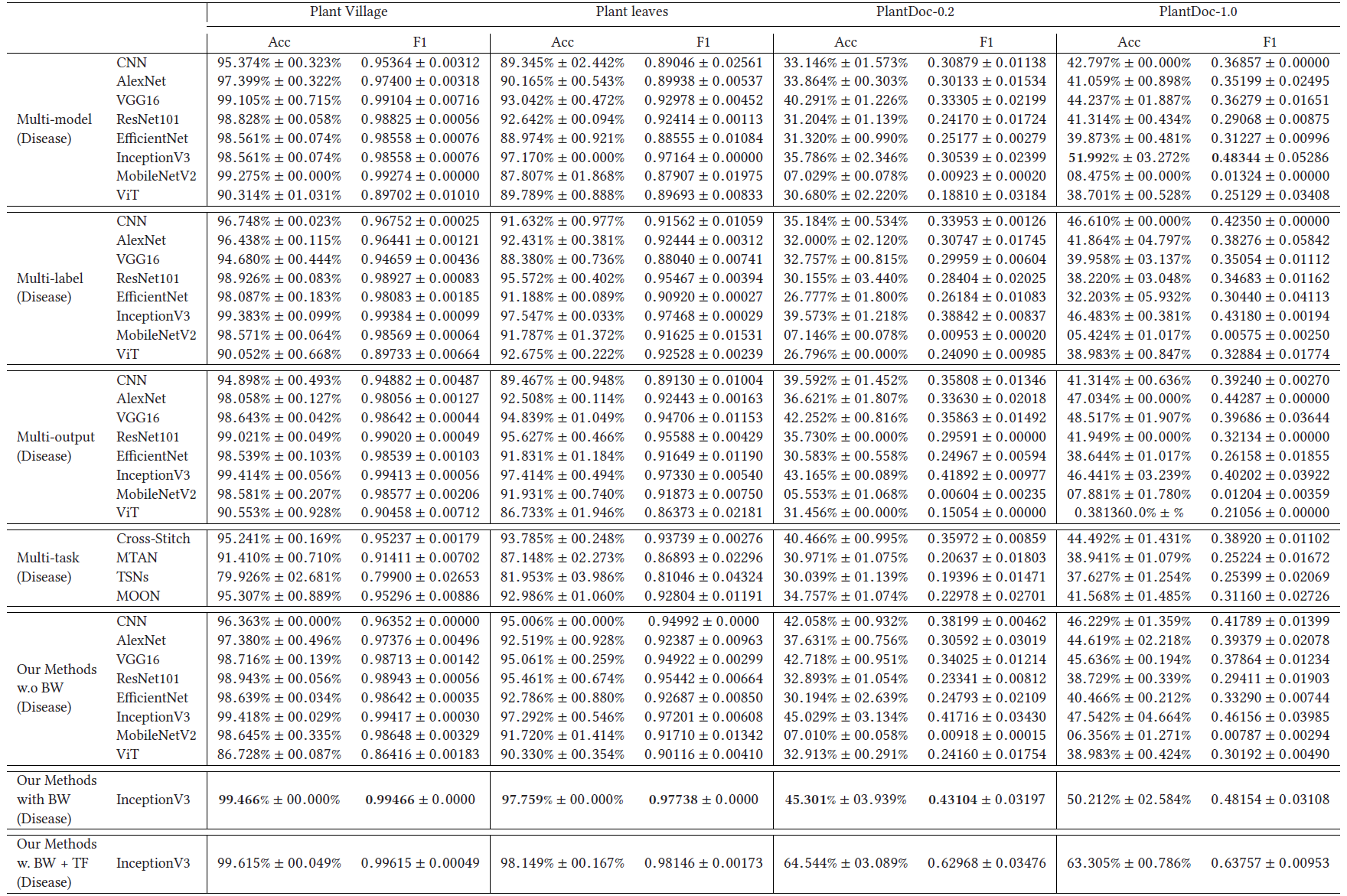
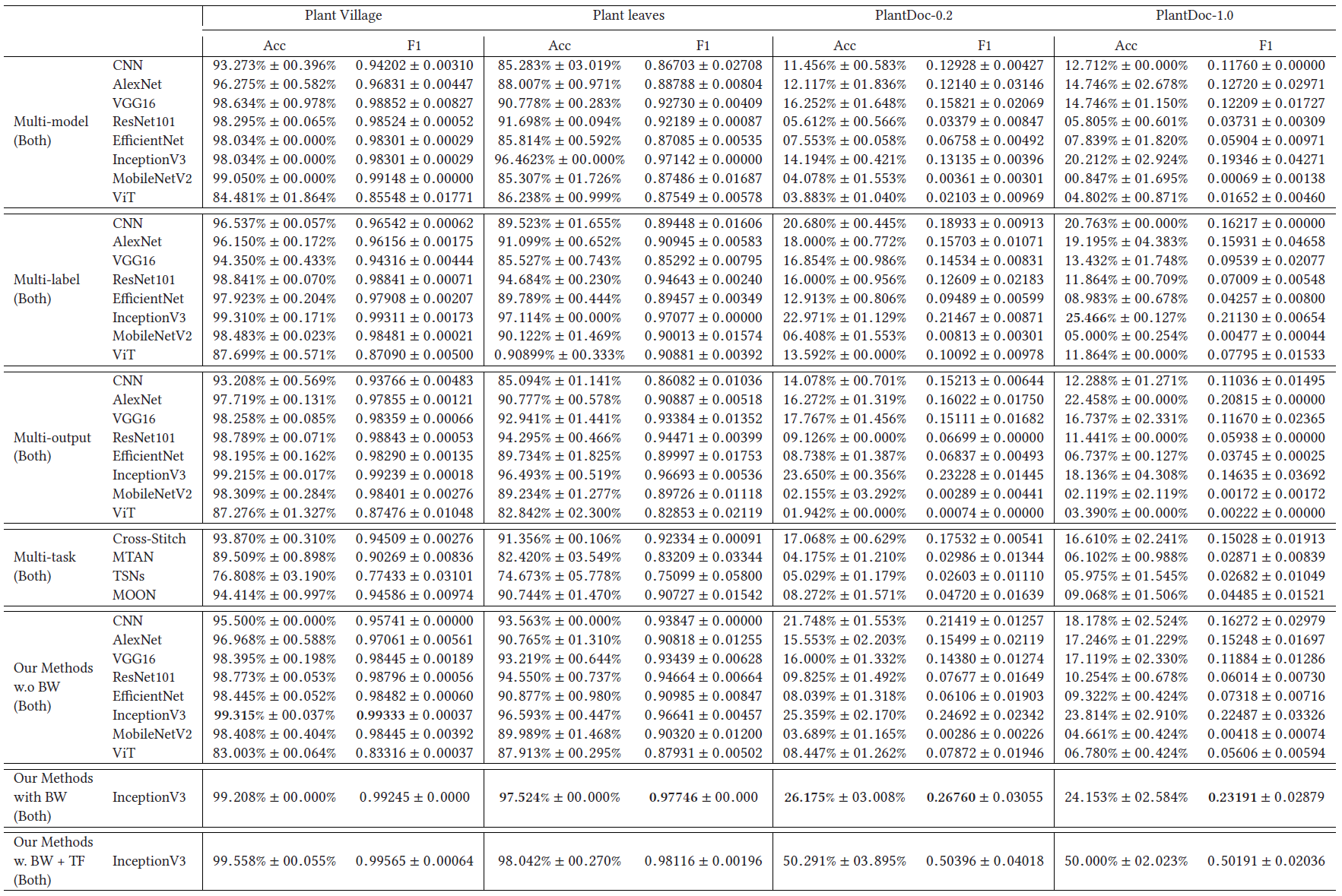
Parts of the comparison & results are as follow. Please see more details in our paper "Deep Learning for Plant Identification and Disease Classification from Leaf Images: Multi-prediction Approaches". Thank you.
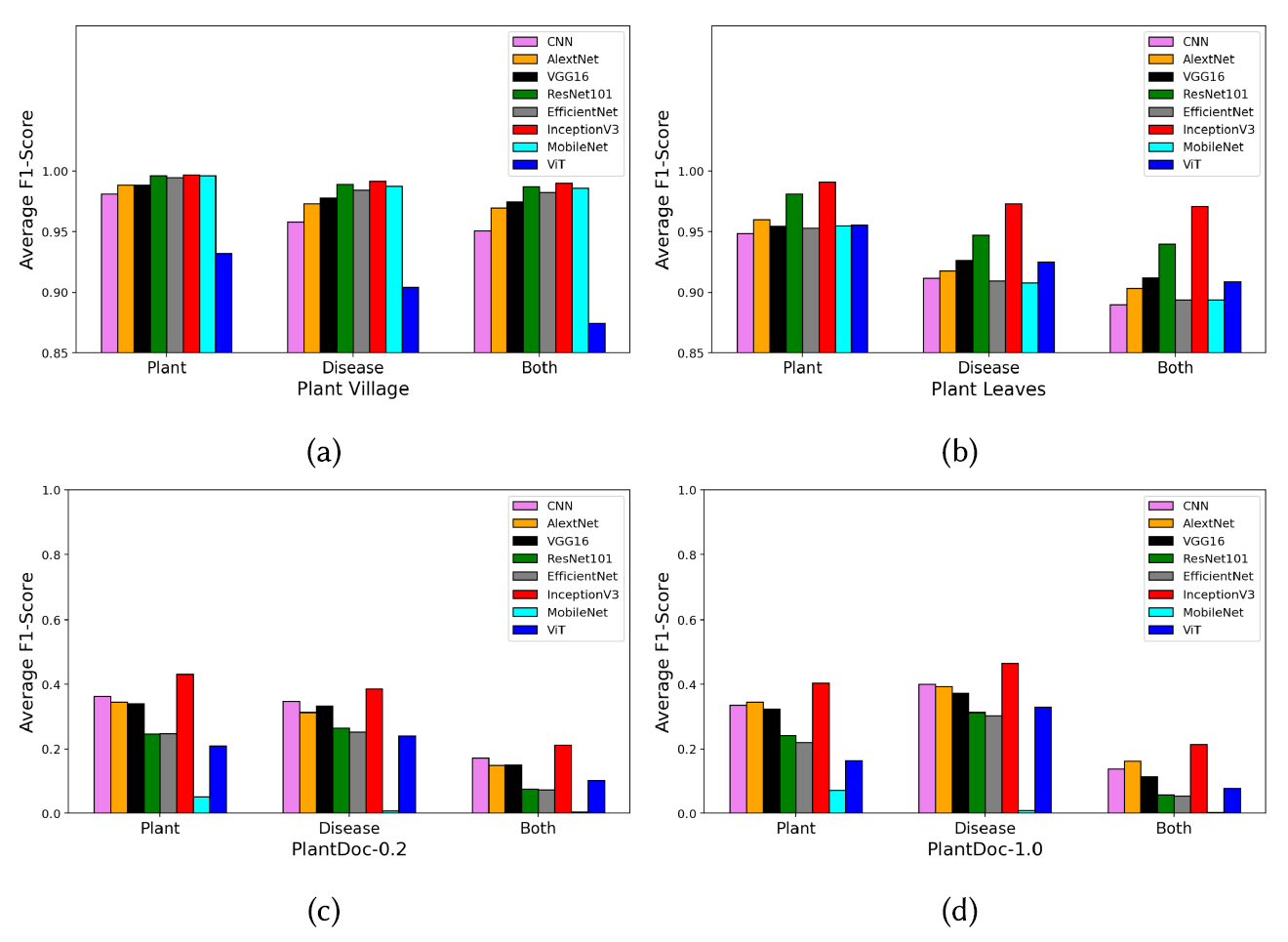
Comparison of backbone CNNs
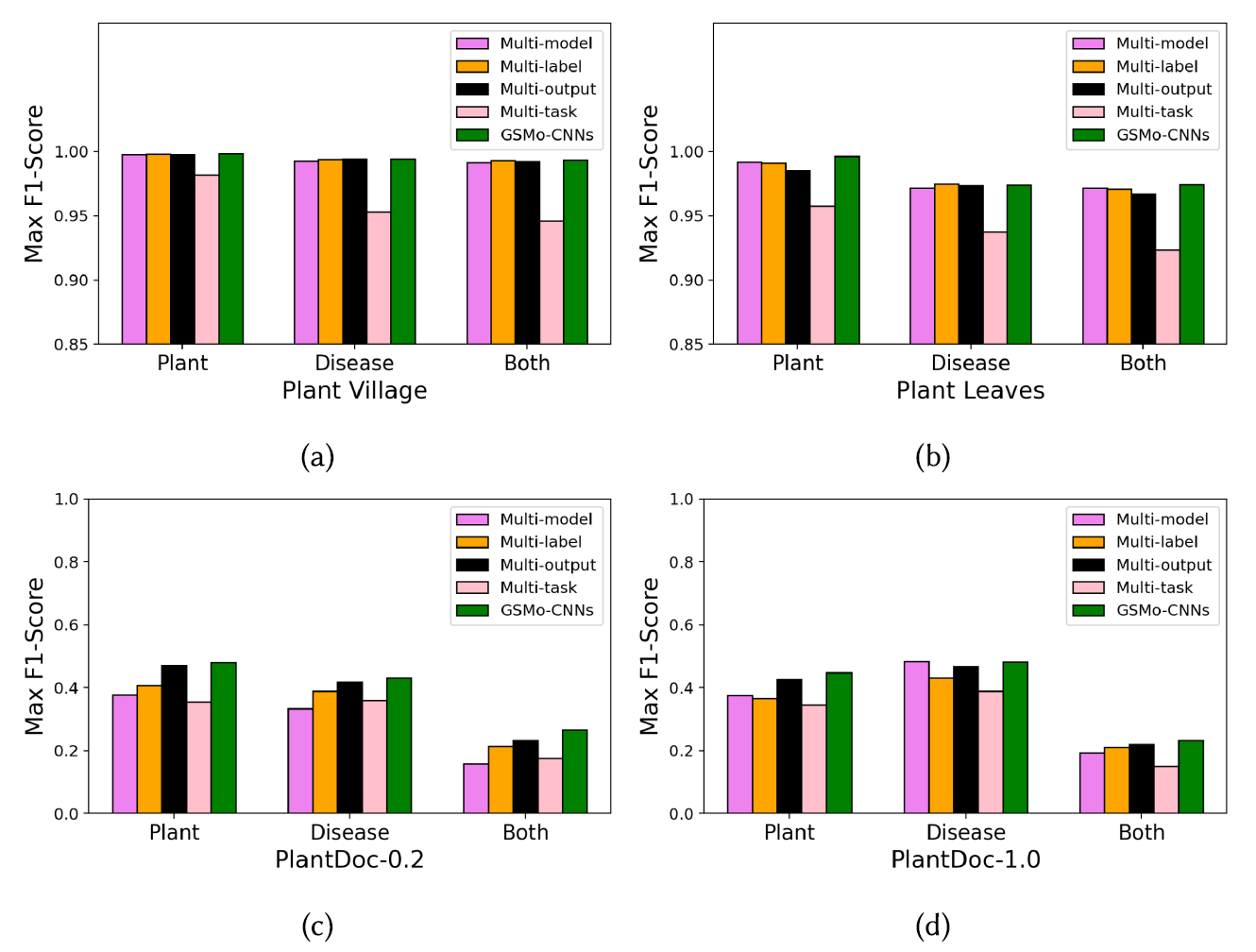
Comparison of approaches
Plant Type Results
Plant Disease Results
Both (Type & Disease) Results