Brivez is a bioinformatic tool thought as Quality of Life's improvement, providing high quantity of data in a snap, giving you a quick view on what you could find inside your transcriptome/sequences' list.
Installation requirements aside, you will need two files:
domain_profile.hmm
It goes inside --> 01_hmm_profiles foldersequence(s).fasta
It goes inside --> 02_fasta_target
Suggested way to proceed:
- Read the documentation
- The first time follow the checklist
- Run ./brivez_main.sh
At the moment this program runs exclusively on Linux (tested on Debian 11 and Ubuntu 22.04).
See future updates for Mac and Windows.
All the software used are OpenSource.
| Brivez runs locally |
|---|
|
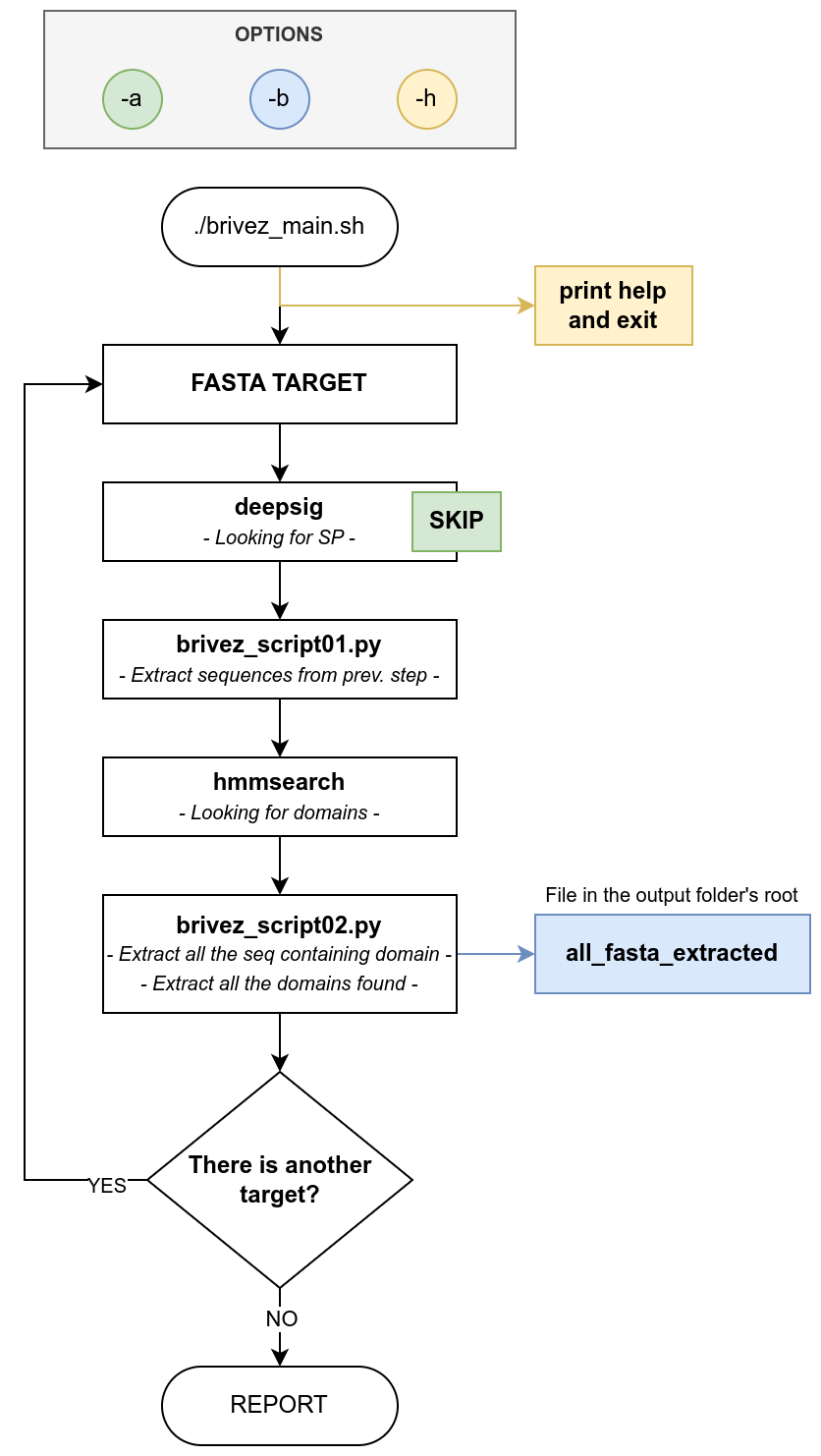
Brivez analyses every fasta file insiede 02_fasta_target.
Every single run follow these workflow:
- Runs Deepsig looking for Signal Peptide (can be disabled with
-a) - Uses the sequences found in the previously step
- Searches for domains using the hmm file inside
01_hmm_profiles- If Brivez founds more than one hmm file, it asks which one choose
- Generates multiple output files for each fasta target
- A file with all the domain extraced can be genereted with
-boption
- A file with all the domain extraced can be genereted with
Flowchart realized with to draw.io_
- Conda (minimum ~3 GB)
- Environment inside Conda with:
- Deepsig (~50 MB) --> needs PYTHON 3.8 !
- Pandas (~15 MB)
- Bio-conda (channel)
- fnmatch (samtools ~1 MB)
- HMMER3 v3.3.2 (~20 MB)
- Brivez (~1 MB)
01 - Quick install for Conda (following the online doc is suggested):
- Download the installer at this link
- Verify your installer hashes
- In the terminal run
bash Anaconda-latest-Linux-x86_64.sh
Create an environment ad hoc
conda create -n bioinfo-brivez python=3.8
To see all the env created
conda env list - Choose the environment with
conda activate bioinfo-brivez
to close it use
conda deactivate bioinfo-brivez
---- By this point be sure to have activated the right environment! ----
02 - Install in Conda some stuff:
conda config --add channels biocondaconda install -c bioconda samtoolsconda install -c conda-forge dpathconda install pandas
03 - Install the predictor of signal peptides, DeepSig
- Just as described on its site, use
conda install -c bioconda deepsig
04 - Install HMMER3
- Just as described on its site, use
sudo apt-get install hmmer
(v3.3.2 both on Ubuntu 22.04 and Debian 11)
Otherwise is installable following the official documentation
05 - Download Brivez
- Download the Brivez folder with
git clone https://github.com/furacca/brivez
or whatever way you prefer
-
01_hmm_profiles- contains your hmm files (check hmm - Pfam download)
-
02_fasta_target- contains your fasta files
- the only accepted format is fasta (no fas, fa, ...)
-
Be sure to have the right environment activated
conda activate therightenvironment
-
Run Brivez with
./brivez_main.sh
Inside the 03_results you will have your output folder, all with the same structure:
- Research_number_XXX
- fasta_target1 (directory)
- multiples files
- fasta targetX (directory)
- multiples files
- Report (file)
- fasta_target1 (directory)
For every fasta target there will be one folder containings the following:
- 01_deepsig-analysis.tsv
- Deepsig analysis; if
-ais enabled, then is empty
- Deepsig analysis; if
- 02_sequences_selected.fasta
- Sequences with SP by deepsig; if
-ais enabled, all sequences are selected
- Sequences with SP by deepsig; if
- 03_hmmsearch_output_table
- hmmsearch output
- 04_hmmsearch_output_table_data_parsed
- hmmsearch output parsed in csv
- 05_pandas_sequences_table.tsv
- pandas table with the list of all domains found
- 06_sequences_which_have_domains.fasta
- list of sequences presenting domains found
- 07_extracted_domains.fasta
- domains found extracted by the selected sequences
- 08_extracted_domains.csv
- same as 07, but in csv format
- 09_domains_found.csv
Important The 08_extracted_domains.csv file can be easyly read in calc/excel selecting all the cells and clicking on Data -> Autofilters.
- hmm - Pfam download
Contains tutorials on:- How to download the full Pfam database
- How to download a specific ~.hmm file from pfam
- Hmmer cheatsheet
None (yet).
TOP PRIORITY
- None
MEDIUM PRIORITY
- Simpler way to user Brivez (Docker? Appimage?)
- List of the sequences with just one domain type (needing full Pfam database)
LOW PRIORITY
- Check some solution for Mac and Windows (Docker?)
- Extracted list of all SP