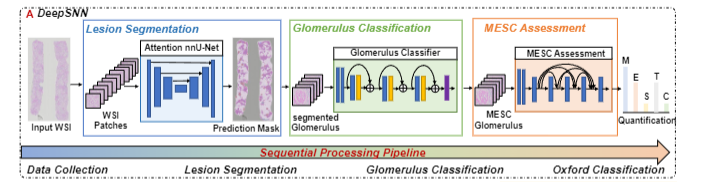
A large-scale dataset and a deep sequential neural network for IgA nephropathy histopathology identification and quantification
- This is the official repository of our paper entitled "A large-scale dataset and a deep sequential neural network for IgA nephropathy histopathology identification and quantification".
1. Environment
Python 3.7
PyTorch = 1.5.0
opencv-python
You can install them by the requirements.txt
pip install -r requirements.txt2. Dataset
The dataset will be uploaded to Baidu Disk (LINK:https://pan.baidu.com/s/1uH9qmjYKhGvIOPjiXv2v_A). You should ask for the PASSWORD to download it by sending emails to pansai301@126.com. The email should contain your institution, your grade/occupation and the purpose of pursuing this dataset. The act of spreading our dataset freely on the Internet is forbidden.
3. Model
You can Download the pytorch model from BaiduPan(key: BuAa) and put the .pth files in the repository folder.
4. Test
-
Clone this repository:
https://github.com/fyb99/DeepSNN.git -
Place the test images in
./data/. For example,./data/test_sample.data └── test_sample ├── img.png └── mask -
Run the following command for the IgA nephropathy histopathology identification task with the input whole slide image.
python isolate_test.py -
Finally, you can find the predicted results in
./data/test_sample/predicted.
5. Training
- If you want to train the proposed method with our dataset or private dataset. Run the following command for training the lesioin segmentation subnet.
python vanilla_train_seg.py - Run the following command for training the glomerulus classification subnet.
Note how the training sets of two subnets are generated can be found in
python vanilla_cls.pydataset.pyandcls_dataset.py.