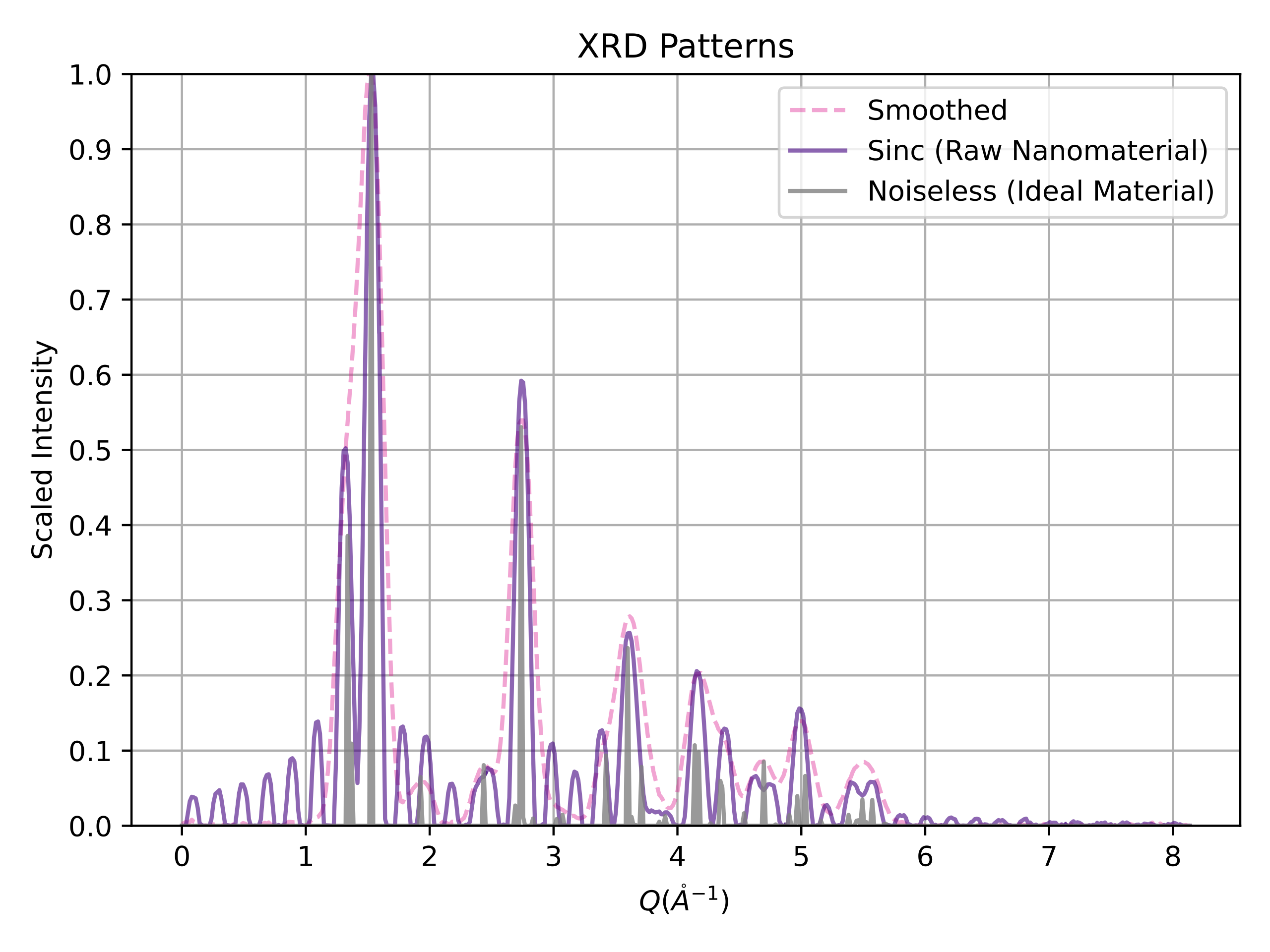
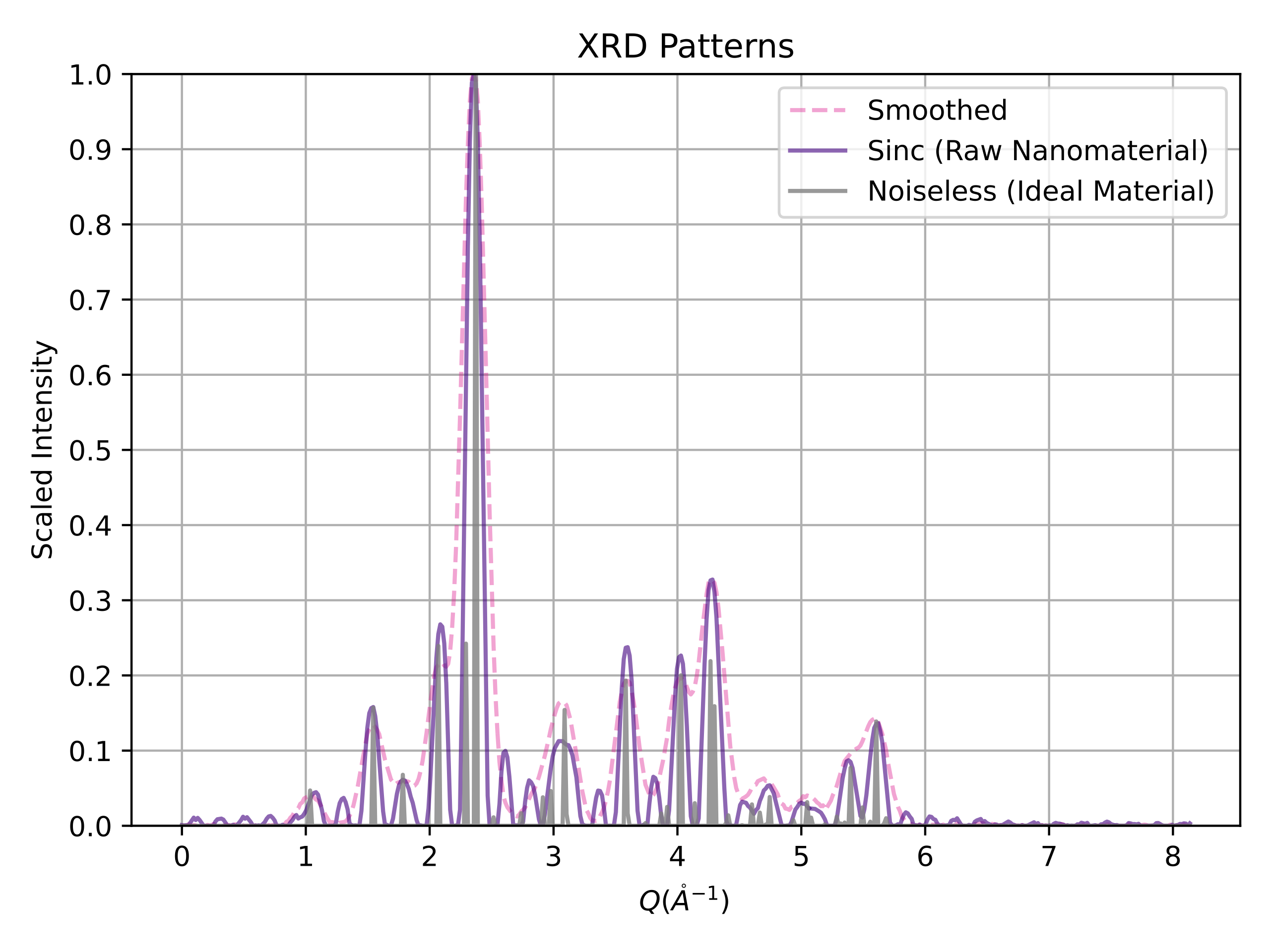
This is the repository for XRDnet, the world's first end-to-end nanostructure solver from powder x-ray diffraction (PXRD) patterns. Associated article: Ab Initio Structure Solutions from Nanocrystalline Powder Diffraction Data via Deep Generative Modeling.
All code blocks assume you start from this directory.
Much thanks to CDVAE.
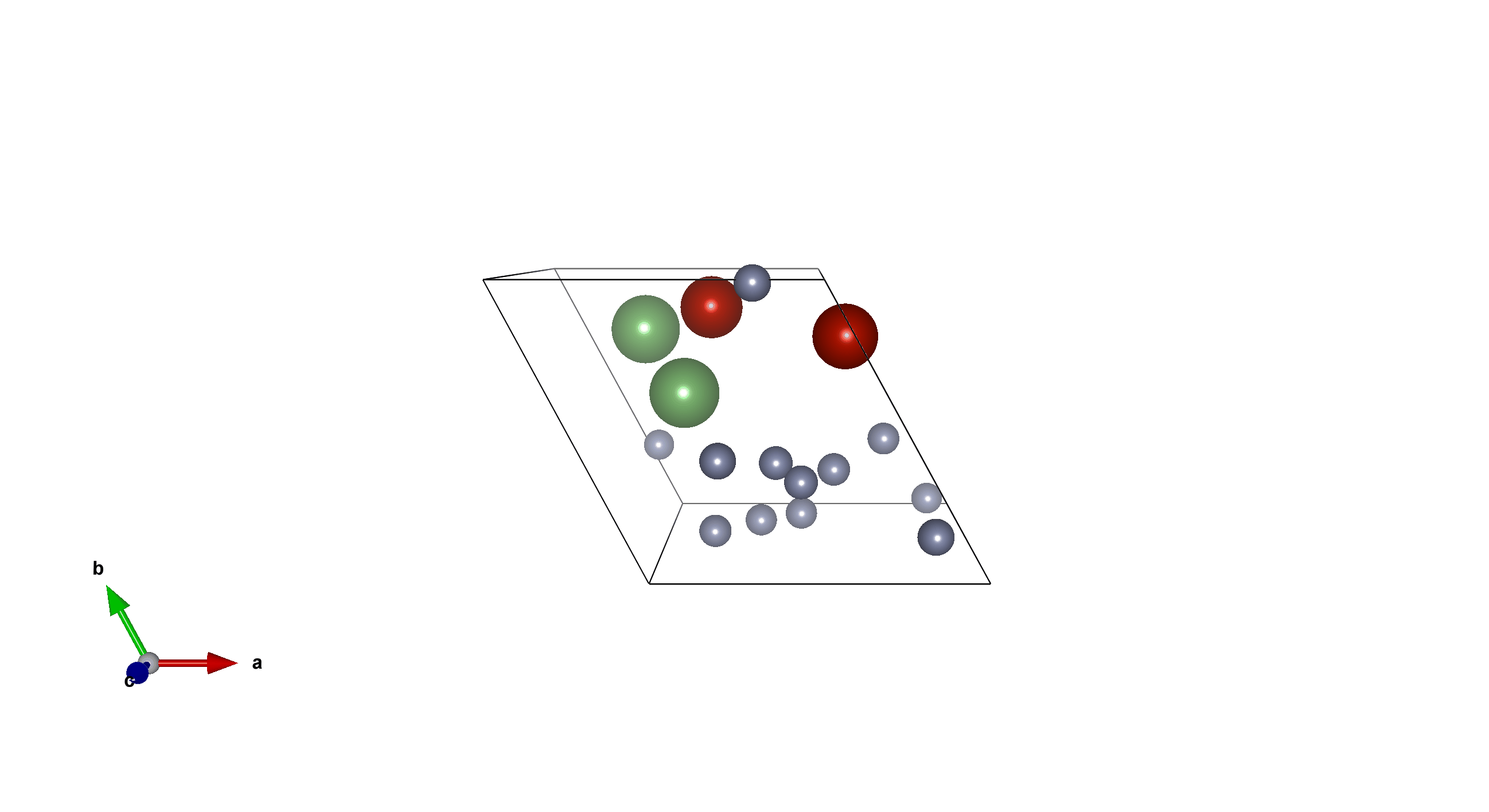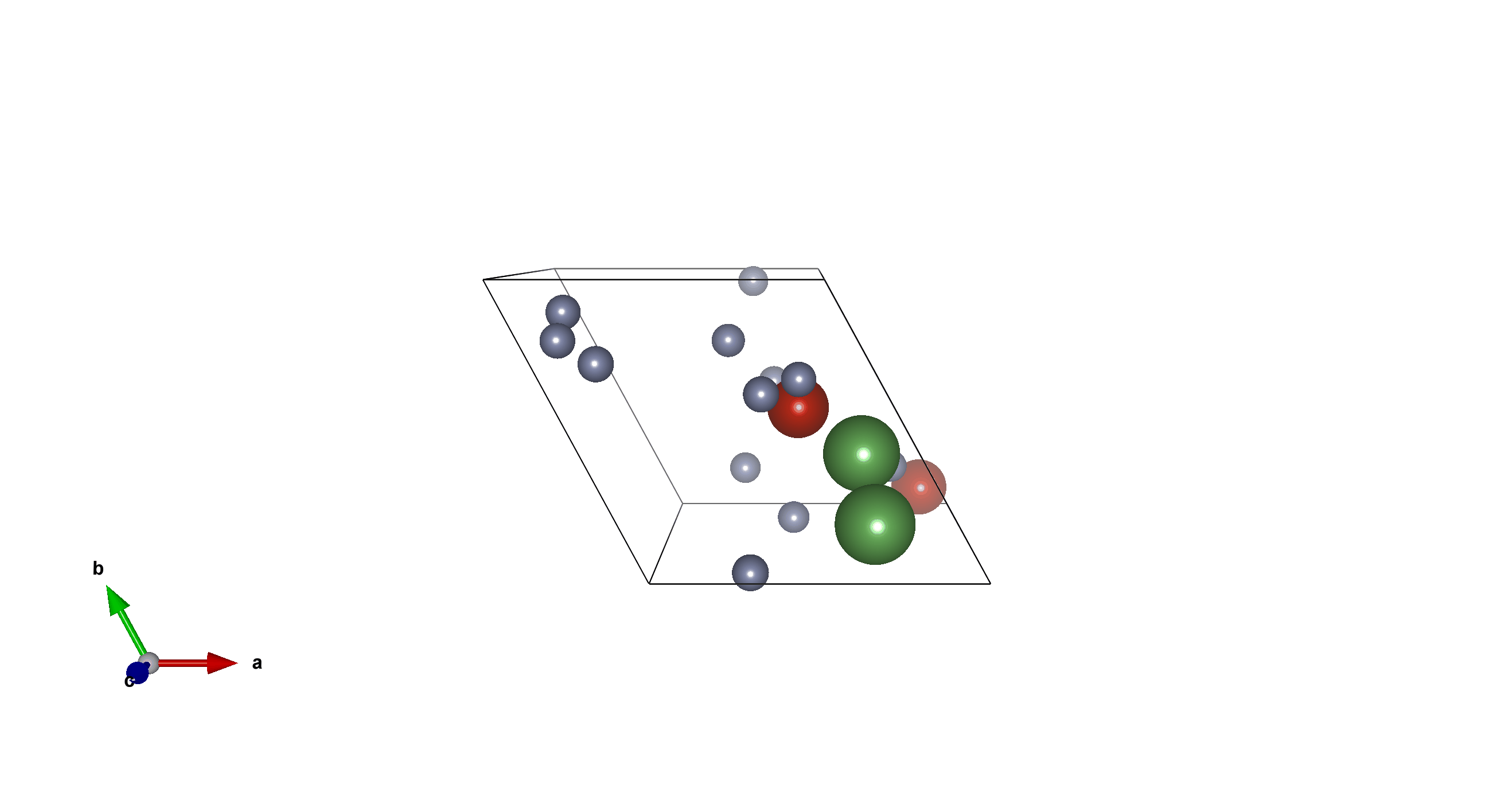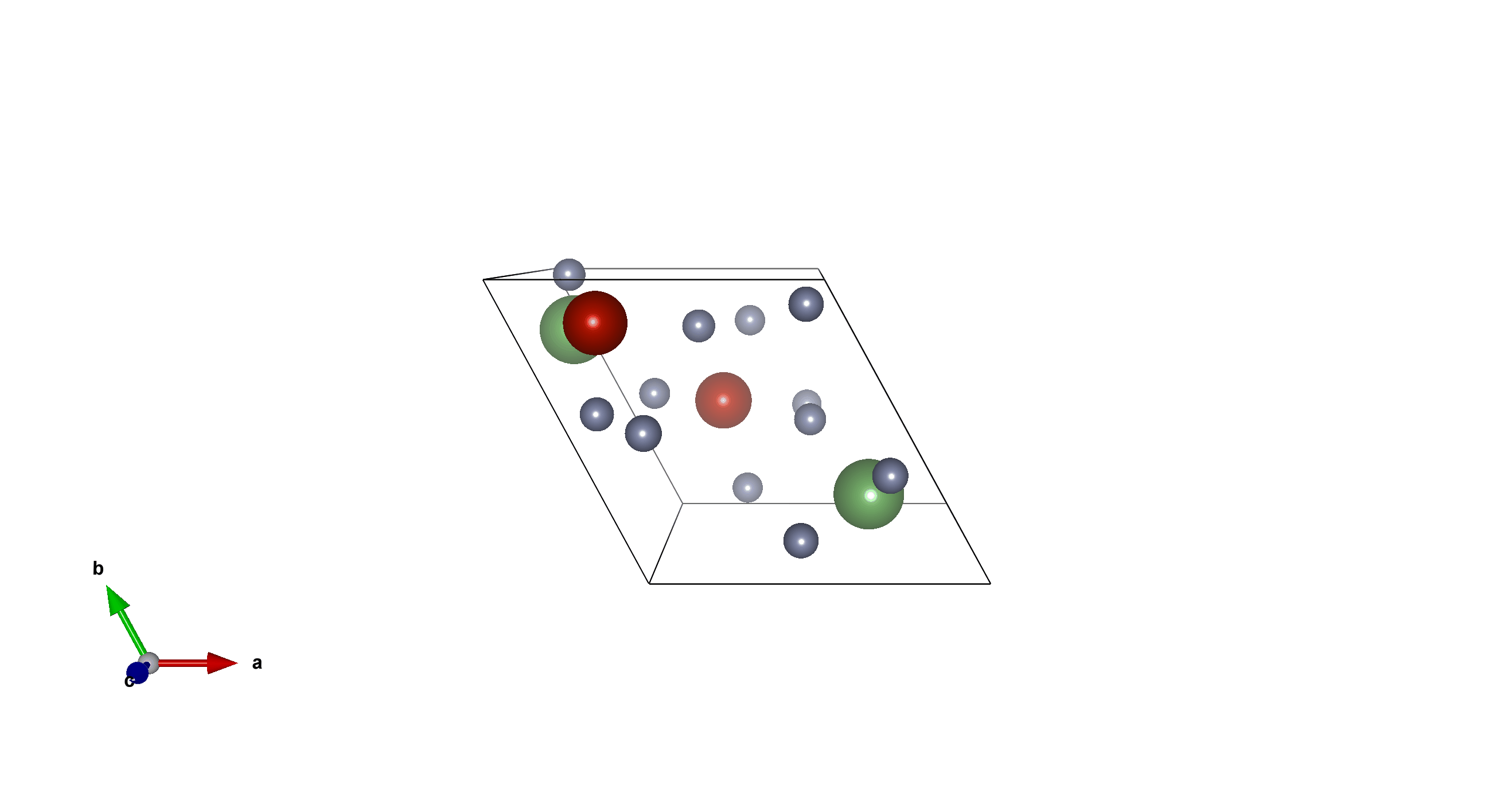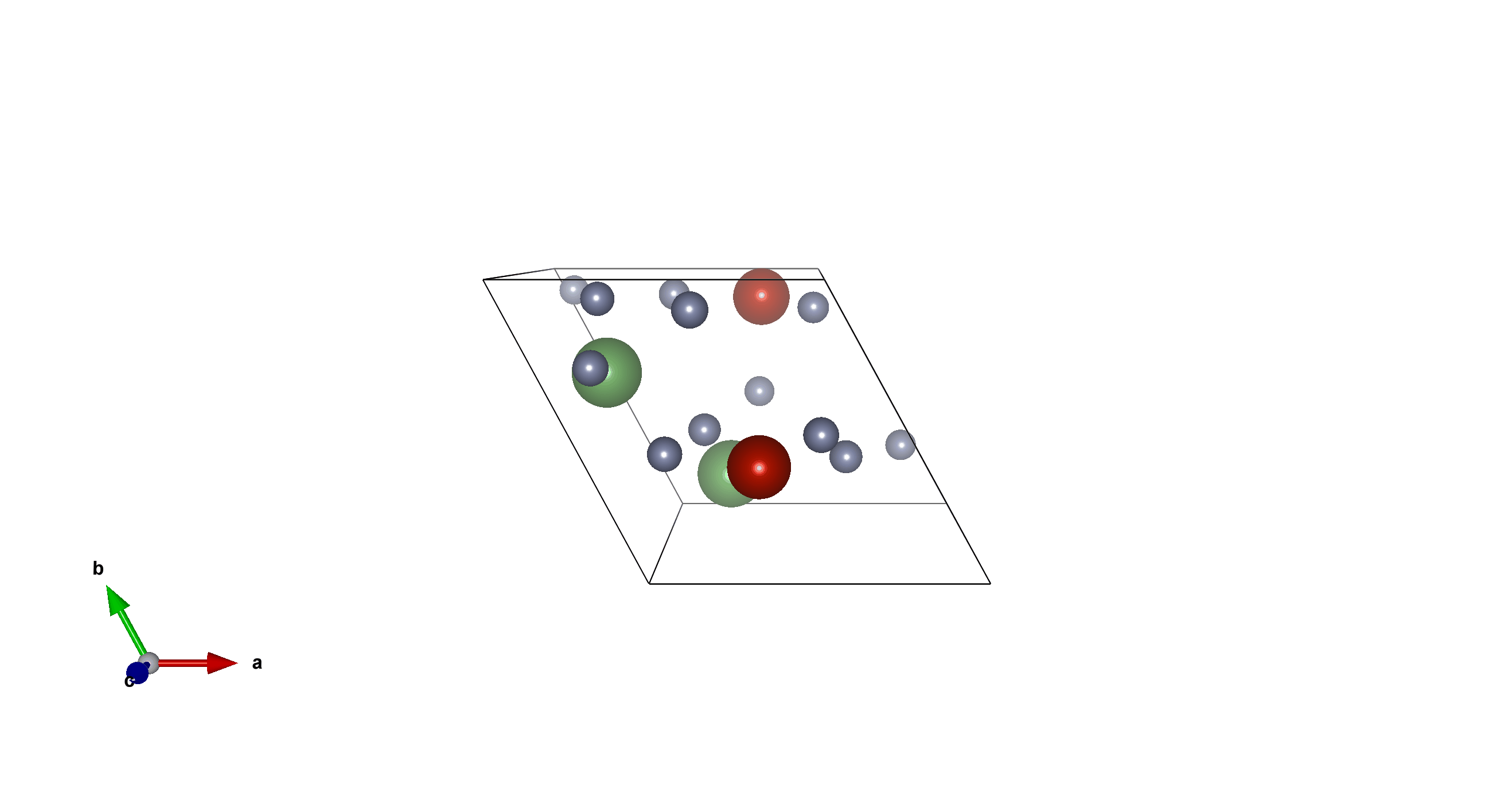
Li2V2F12
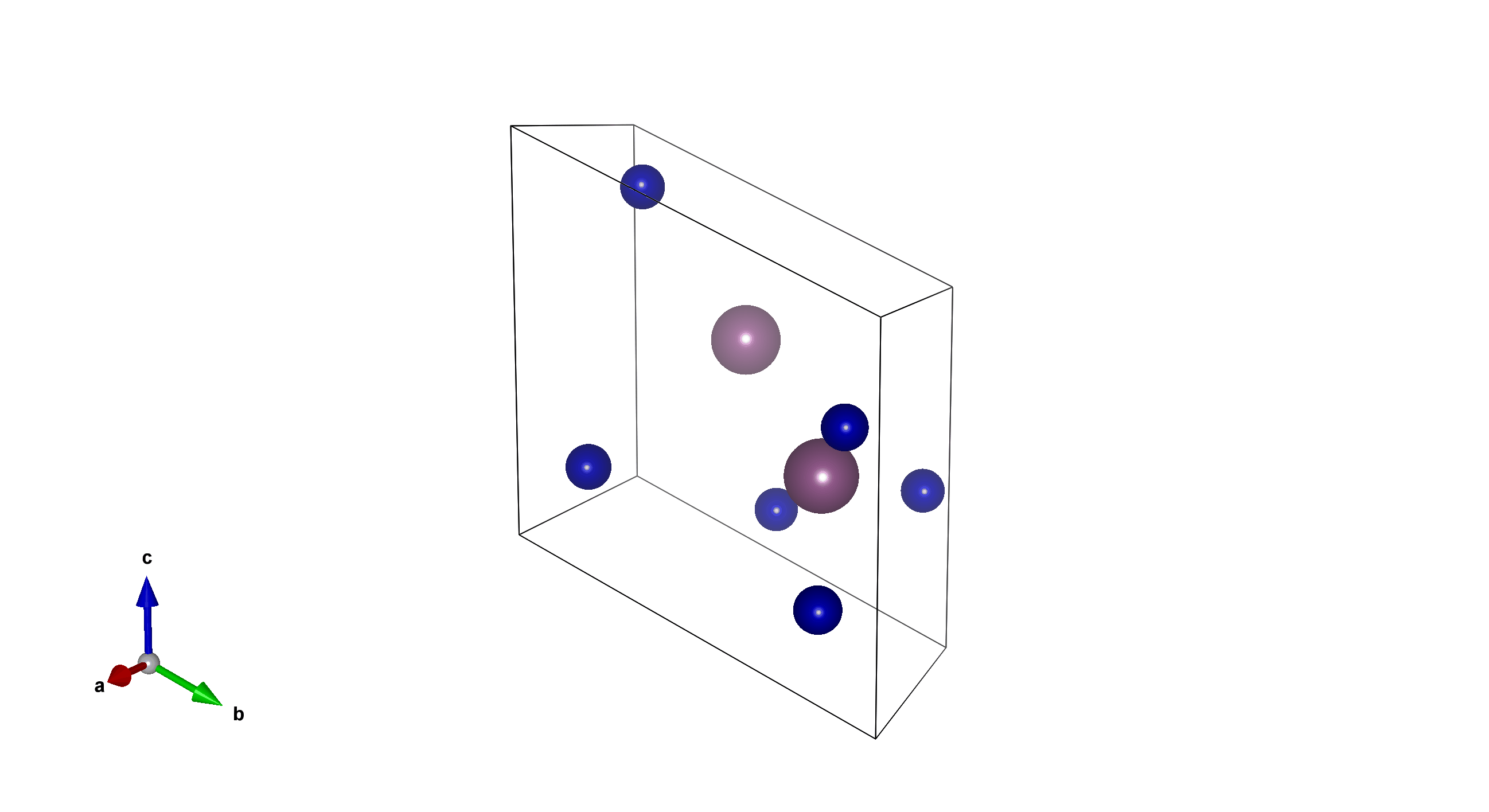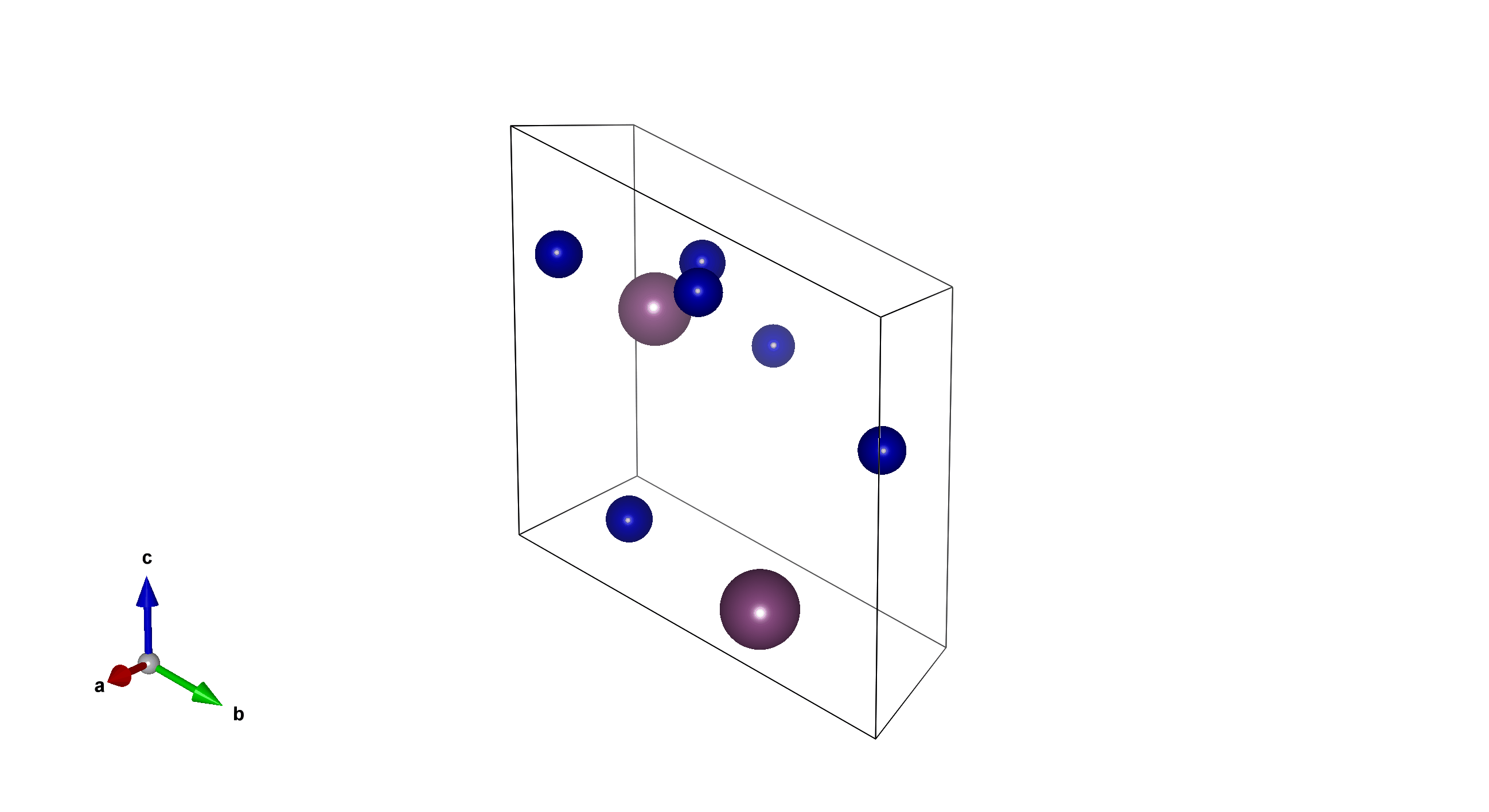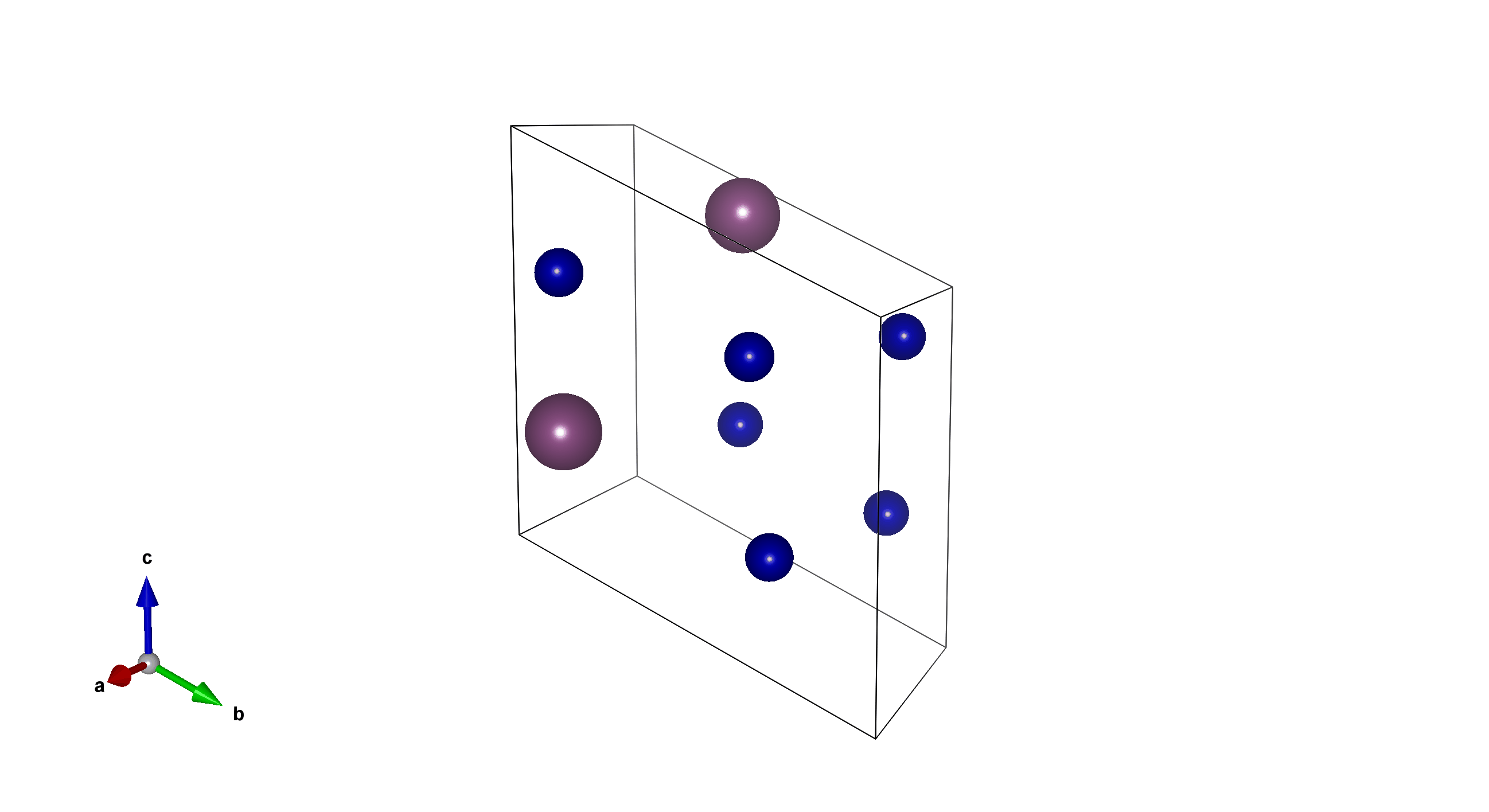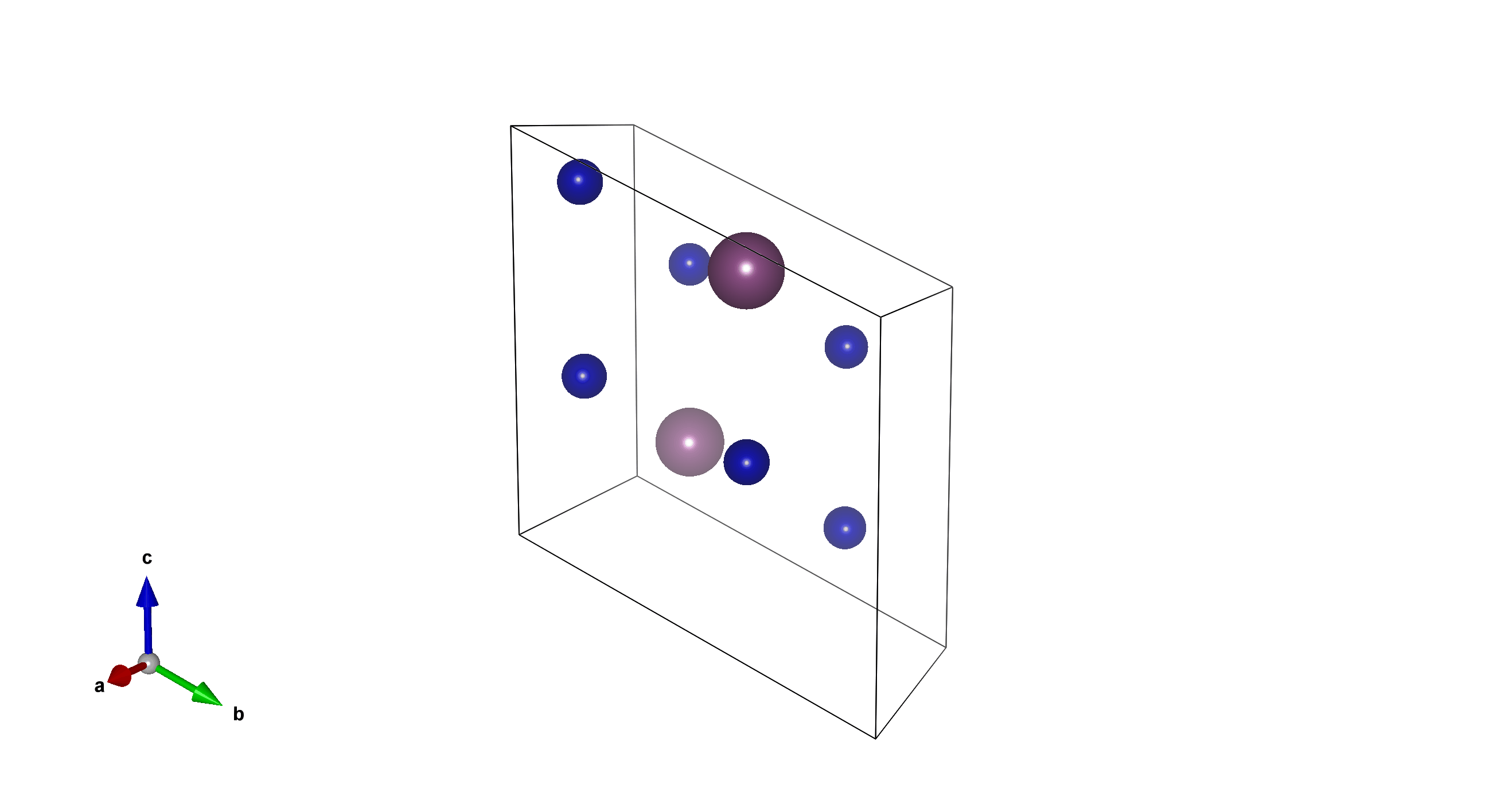
Tm6Sc2
Use Python 3.9.18 with Linux.
In our experience, depending on the system, you may have trouble with:
If so, just follow the instructions on their GitHub repos to install the versions that align with your CUDA version. Here are the suggested ways to do so:
pip install torch==2.0.0
pip install torch-scatter -f https://data.pyg.org/whl/torch-2.0.0+${CUDA}.html
pip install torch-sparse -f https://data.pyg.org/whl/torch-2.0.0+${CUDA}.html
After that, run the following command to install remaining requirements.
pip install -r requirements.txt
Setup environment variables by making a copy of the .env.template file and rename it to .env. Modify the following environment variables in .env.
PROJECT_ROOT: path to the folder that contains this repoHYDRA_JOBS: path to a folder to store hydra outputsWABDB: path to a folder to store wabdb outputs
Finally, install this package with
pip install -e .
This process, if done correctly, should take less than an hour.
See data/mp_20 for MP-20-PXRD, our modification of MP-20 with PXRD patterns (no broadening added).
See data/experimental_data for instructions on obtaining the crystal structures with experimentally observed PXRD patterns (we do not own them; IUCr does).
Despite the filenames, they're not .csv files (they're pickle files), because the data contains PyTorch tensors.
No other action needs to be taken on data, as the scripts automatically load the datasets.
This is unnecessary for MP-20, because we already have created and uploaded the datasets for you. However, if you wish to try something new on the MP-20 dataset (let's say, different wavelengths or data splits), you can follow these instructions.
Before running, replace /home/gabeguo/ in create_data.sh with your home directory. This takes less than an hour.
cd scripts
bash create_data.sh
Data should be saved in data/mp_20.
This trains the ../hydra/singlerun/[today's date].
On a single GeForce RTX 3090 (24 GB), each model should take about one day to train.
cd scripts
CUDA_VISIBLE_DEVICES=x bash train_mp20_model_sinc10.sh
CUDA_VISIBLE_DEVICES=x bash train_mp20_model_sinc100.sh
You will have to change --model_path inside each script to have the appropriate home directory (rather than /home/gabeguo/) and date (rather than 2024-04-07).
On a single GeForce RTX 3090 (24 GB), each evaluation (per model) should take about one day to conduct.
cd scripts
CUDA_VISIBLE_DEVICES=x bash conditional_generation_sinc10.sh
CUDA_VISIBLE_DEVICES=x bash conditional_generation_sinc100.sh
cd scripts
CUDA_VISIBLE_DEVICES=x bash conditional_generation_random_baseline_sinc10.sh
CUDA_VISIBLE_DEVICES=x bash conditional_generation_random_baseline_sinc100.sh
cd scripts
CUDA_VISIBLE_DEVICES=x bash conditional_generation_sinc10_baseline_noOpt.sh
CUDA_VISIBLE_DEVICES=x bash conditional_generation_sinc100_baseline_noOpt.sh
Your file directory should look something like this:
cdvae_xrd/
... [some stuff here] ...
hydra/singlerun/
[whatever date you trained model on]/
mp_20_sinc10/
.hydra/
config.yaml
hydra.yaml
overrides.yaml
hparams.yaml
... [other stuff here] ...
mp_20_sinc100/
... [same stuff here] ...
Run the following code (assuming you are in cdvae_xrd) to create the proper evaluation setup for experimental data:
cd ../hydra/singlerun/[whatever date you trained model on]
cp mp_20_sinc10 mp_20_sinc10_EXPERIMENTAL_TEST
Now, go into mp_20_sinc10_EXPERIMENTAL_TEST/.hydra/config.yaml and change line 7 to be:
root_path: ${oc.env:PROJECT_ROOT}/data/experimental_xrd
from
root_path: ${oc.env:PROJECT_ROOT}/data/mp_20
Do exactly the same change for mp_20_sinc10_experimental/hparams.yaml.
Again, remember to change --model_path inside each script to have the appropriate home directory (rather than /home/gabeguo/) and date (rather than 2024-04-07).
This should only take a few hours at most, due to there being fewer experimental PXRD patterns.
cd scripts
CUDA_VISIBLE_DEVICES=x bash conditional_generation_experimental.sh
CUDA_VISIBLE_DEVICES=x bash conditional_generation_baseline_noOpt.sh
CUDA_VISIBLE_DEVICES=x bash conditional_generation_random_baseline_experimental.sh
As before, in the __main__ part, change the home directory from /home/gabeguo/ to whatever your home directory is.
This should take less than an hour.
cd scripts
python calculate_xrd_patterns_post_hoc.py
python calculate_r_factor_post_hoc.py
bash calc_r_value_distribution.sh
Reiterating (as you've already guessed), in extract_results_by_crystal_system.sh, change the home directory from /home/gabeguo/ to whatever your home directory is.
This should take less than an hour.
cd scripts
bash extract_results_by_crystal_system.sh
If you find the code in this repository helpful, please cite the following:
@article{guo2024diffusion,
title={Diffusion Models Are Promising for Ab Initio Structure Solutions from Nanocrystalline Powder Diffraction Data},
author={Guo, Gabe and Saidi, Tristan and Terban, Maxwell and Billinge, Simon JL and Lipson, Hod},
journal={arXiv preprint arXiv:2406.10796},
year={2024}
}