version: 3.0.0
If OONI were a car, the OONI Pipeline would be it's engine
Pipeline is responsible for aggregating data from network measurement clients (such as OONI Probe) and analyze it to identify network anomalies.
The main goals are:
-
Have a reasonable end-to-end latency (the delta from when a measurement is uploaded to when it appears in api.ooni.io). Our target is minutes/hours, not seconds.
-
Make it easy to re-run new heuristics on top of the existing dataset
-
Ease parallelization of CPU intensive tasks to use all available cores on a single machine
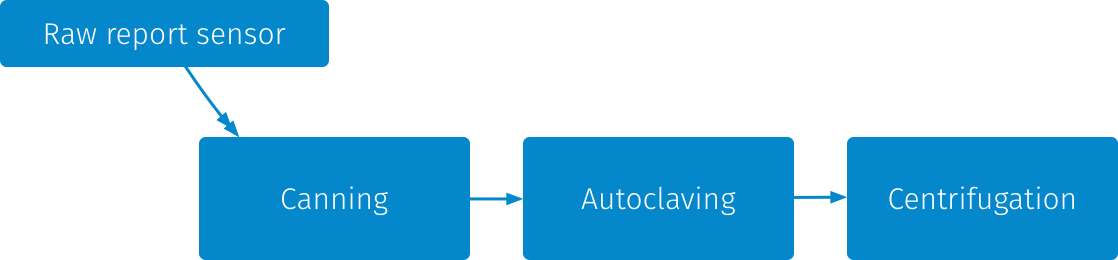
The OONI Pipeline is currently based on primarily 3 stages, that are run on every "tick": 1. Canning, 2. Autoclaving, 3. Centrifugation.
The purpose of the canning stage is to convert report files, which are collections of measurements, into a more compact representation that makes the files occupy less space on disk, but also allows us to reprocess the measurements faster.
When we intend to re-run the heuristics against historical data, this stage is going to be skipped most of the times.
This is done in the following way:
canning-1.py | tar c 2000-13-42/foo.yaml 2000-13-42/bar.json
| canning-2.py | lz4 -5
| canning-3.py
canning-1.py: groups reports by test_name, slices them into ~64 Mbytes groups of
uncompressed data and send them to tar.
canning-2.py: reads a .tar file, verifies the sizes of reports, calculates
the sha1 and crc32 of each report, calculate size of tar file and then pipes
the unmodified tar file to lz4.
canning-3.py: reads a .lz4 from the pipe, calculates it's size, sha1 and
crc32, dumps the lz4 file & all the checksums to disk.
Note: the script names are not the names of the actual script files, but are just used for example purposes.
All the sizes are recorded to be checked to avoid various sort of unexpected file truncations.
The purpose of the autoclaving stage is to perform normalization and sanitization of the report files including PII cleanup if leak is discovered. This means converting legacy YAML reports to JSON as well as converting all measurements to a consistent JSON format (and performing some fixes to the data format to avoid surprising consumers of the data).
In other words, it converts tar.lz4 files in a 1:1 way.
lz4 compression is a bit tricky at this stage: lz4 frames are independent,
report offsets within the block are recorded in the separate index file, so the
resulting file is both streamable (with ability to skip tar records without
decoding), seekable (single lz4 frame is read to get a single report — that's
~56kb of compressed data) and readable with ordinary tar -I lz4 --extract for
whoever wants to parse sanitized measurements.
Indexes for the reports are stored as well, they include block offsets, sizes, and offsets within block required to seek() to exact measurement and to read the file in streaming way without parsing tarfile stream.
Centrifugation is where the interesting things start happening. It's what is done to "enrich" the data. It reads data sanitized by autoclaving, extracts the essence that can be aggregated and stores metadata into a PostgreSQL database for further processing.
Note: in the airflow DAG view this is actually called meta_pg.
This is not technically a task, but is rather a "sensor" in airflow lingo. What
this means is that a check is done at chameleon (the host that aggregates all
measurements from OONI collectors) to see if there is more work to do and if so
will start a new iteration of the DAG.
The technological stack of the pipeline is the following:
- Airflow as a workflow manager or put otherwise a "enhanced cron"
- PostgreSQL as the primary database used to store metadata
- Docker to create containers for the various stages of the pipeline, without having to worry about dependency management
- Ansible to make deployment automation easier
In this section we will go over some of the common tasks related to operating and contributing to the OONI Pipeline.
It's important to keep in mind that every task inside of the Airflow DAG needs to be optimized for big units of computation, you should avoid having small tasks, otherwise you incur in a lot of scheduling/orchestration overhead.
Most of the time you will probably care to edit the centrifugation stage instead of adding a new DAG task, to avoid re-reading the measurements multiple times.
- Edit the centrifugation file
The file to edit is located in af/shovel/centrifugation.py.
Most of the time you will be adding a new Feeder. A Feeder is something
that populates a given table with a particular class of measurements.
To create a new Feeder you will have to implement some methods:
class MyFeeder(BaseFeeder):
min_compat_code_ver = 4
data_table = sink_table = 'my_fancy_test'
columns = ('msm_no', 'some_value')
@staticmethod
def row(msm_no, datum):
ret = ''
if datum['test_name'] == 'my_fancy_test':
some_value = datum['test_keys'].get('some_value', None)
ret = '{:d}\t{}\n'.format(
msm_no,
pg_quote(some_value) # if it's nullable or string
)
return ret
@staticmethod
def pop(datum):
test_keys = datum['test_keys']
test_keys.pop('some_value', None)The row method should return a row in COPY FROM syntax to populate the sink_table with the
extracted metadata.
The pop method should pop all the extracted keys (or the keys specific to this test that are to be ignored), so that the resulting datum is "clean".
The sink_table class attribute defined which table is going to be written and
columns specifies which columns should be present in such table.
data_table defines the table to cleanup in case of reprocessing, that's
useful if sink_table is temporary (in that case you'll likely have to
implement close() method that melds temporary table into persistent one).
min_compat_code_ver defines centrifugration code versions that are
compatible with this extractor. If you are adding a new feeder, then this
should contain the single bumped CODE_VER string (more on this
below).
1b. (optional) Create SQL migration script
If you are populating new tables, or the schema of existing tables need to
change, you need to create a new SQL migration script inside of af/oometa/.
Be sure to write both the install and the rollback version of it.
In the install, wrap everything inside of a transaction and call the
versioning helper, for example:
BEGIN;
select _v.register_patch( '00x-my-new-schema', ARRAY[ '00x-depends-on' ], NULL );
-- Create your tables or make changes
COMMIT;
In the rollback, you should restore the DB to previous state and call the
unregister method of _v:
BEGIN;
select _v.unregister_patch( '00x-my-new-schema' );
-- Drop the table you create or alter the schema to be in the previous state
COMMIT;
- Push a new docker image
You should then build a new docker image by bumping the version number inside of
af/shovel/build.
The image should then be pushed to dockerhub by running:
docker push openobservatory/shovel:latest && docker push openobservatory/shovel:0.0.NN
- Redeploy an update
To redeploy the updated version of the pipeline you should use ooni-sysadmin.
Bump the version number inside of ansible/roles/airflow/files/docker-trampoline,
then run from the ansible directory:
./play deploy-pipeline.yml --tags airflow-dags
- Reprocess the data
To re-process the data through the Airflow UI and scheduler, do the following:
-
Got to Tree view
-
Click on the task you want to reprocess
-
Click on "Clear" and select "Past"
-
Click "Done"
-
In the confirmation page click on "OK"
Warning: the airflow scheduler is imperfect and has issues handling too
many DAGs being in running state wasting lots of CPU. Moreover, using
airflow backfill is also
error-prone
as backfill process 1) does not go through scheduler so it does not respect
Pools; 2) may delay processing of new buckets for some days because it consumes
all worker slots; 3) marks touched DagRuns as backfilled with magic backfill_
prefix so these DagRuns become always-skipped by scheduler and airflow clear
does not re-trigger them anymore.
In the future we will have a Makefile to run backfilling without airflow
scheduling overhead while preserving task logs and states.
Fingerprints are what we use to detect web censorship and we currently only consider censorship to be "confirmed" when we have a positive ID on the blockpage fingerprint.
In here we explain how you can go about adding new fingerprints to the DB.
- Create the SQL migration scripts
You should use a template the migration file
af/oometa/003-fingerprints.install.sql and
af/oometa/003-fingerprints.rollback.sql.
Based on this create a new set of files in af/oometa/ called
00x-fingerprints.install.sql and 00x-fingerprints.rollback.sql.
- Run the migration script on the DB
You should then manually apply the schema by running the install sql script on
the database directly.
- Bump version information in
centrifugation.py
Look inside of centrifugation.py and bump CODE_VER.
In the same file change min_compat_code_ver in HttpRequestFPFeeder.
- Fix assertion to the fingerprint table
At some point inside of centrifugation.py there is a line that says something like:
assert '916446978a4a86741d0236d19ce7157e' == next(c)[0], 'fingerprint table does not match CODE_VER
You should change the hash value to the output of the following query on the DB:
select md5(
md5(array_agg(fingerprint_no order by fingerprint_no)::text) ||
md5(array_agg(origin_cc order by fingerprint_no)::text) ||
md5(array_agg(body_substr order by fingerprint_no)::text) ||
md5(array_agg(header order by fingerprint_no)::text) ||
md5(array_agg(header_prefix order by fingerprint_no)::text) ||
md5(array_agg(header_value order by fingerprint_no)::text) ||
''
) from fingerprint
- Deploy and re-process the data
Basically you need to do the same thing you did in "Modifying the centrifugation stage" from step 2. onwards.
The task you want to re-run is labelled meta_pg.
Reprocessing the whole dataset takes a couple of days, it's done asynchronously, but the pipeline does not implement any priorities, so it may block data ingestion for a while.
Some examples of tasks that require one or the other:
- extend: fetch data from collectors or backup stuff to S3 after processing
- new dag: fetch data regarding ASNs (as there is no dependency on other tasks)
Also, DAGs may trigger another DAGs, but DAGs are "pausable" so it's another way to reason about extending vs. creation. E.g. one may want to be able to pause processing, but continue ingestion.
You need to edit the DAG inside of ooni-sysadmin repo: https://github.com/TheTorProject/ooni-sysadmin/tree/master/ansible/roles/airflow/files/airflow-dags
Add a BashOperator, get the task_id.
Then add a new switch case inside of
ansible/roles/airflow/files/docker-trampoline.
There are following usual reasons to run partial re-processing:
- PII leaks are identified and corresponding public autoclaved files have to be updated (as well as LZ4 framing in these files), so these files must be re-processed from scratch
- processing schema changes for some "low-volume" test method like
vanilla_tor(~99% of data volume isweb_connectivitytest data) - new files are added to the bucket when pipeline ticks more often than daily
PII leak may be handled with UPDATE autoclaved SET code_ver = 0 WHERE …, 0
is reserved code_ver value also named CODE_VER_REPROCESS.
centrifugation.py will re-ingest alike autoclaved file while preserving msm_no.
It's mostly useful to look at the "Tree view" (ex. http://127.0.0.1:8080/admin/airflow/tree?num_runs=25&dag_id=fetcher) for the DAG, rather than the "Dag
Runs". The failure state can be relative to a full DAG or to a Task.
When in the "Tree View" you can click on a single task and "View Log" to inspect the log that will likely contain the reason for the failure.
Once you have made the necessary fixes that you believe should fix the DAG
(generally this will involve running the ooni/sysadmin playbook
deploy-pipeline), you should "Clear" the state of the task to have it re-run.
When the measurement processing tasks get stuck, pipeline will accumulate a backlog of unprocessed measurements. These unprocessed measurements will end up in the next bucket after it has been unstuck. To signal to consumers explicitly that these buckets are empty, we need to create an "empty bucket" (e.g. new buckets were not created for a while due to temporary pipeline stall or lack of data in the past)
This state looks like the following in the airflow view:
This is done in the following way:
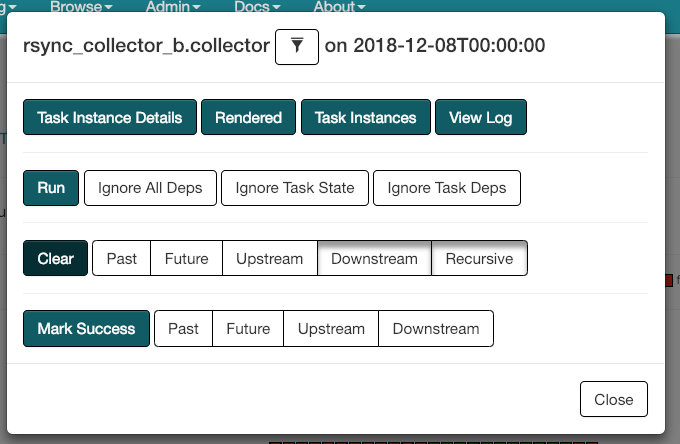
$ ssh datacollector.infra.ooni.io$ cd /data/ooni/private/reports-raw-shals$ sha256sum </dev/null | sudo -u benchmark dd of=YYYY-MM-DD(this will lead to a file withe3b0c44298fc1c149afbf4c8996fb92427ae41e4649b934ca495991b7852b855 -)- Clear state of failed
reports_raw_sensor(the default Downstream & Recursive should be fine) for corresponding date inhist_canningDAG Tree View (you will see two red failed boxes in this view, but it's actually just one task).
The reports-raw-shals file is a "seal" that signals that all the data files
from the collectors are successfully merged into single bucket directory.
It is generated by docker-trampoline script at reports_raw_merge step.
To access airflow web-interface you need to setup a SSH tunnel. It runs at
datacollector.infra.ooni.io.
This line is what you want inside of your config:
Host datacollector.infra.ooni.io
LocalForward localhost:8080 172.26.43.254:8080
More documentation can be found in docs/pipeline-16.10.md