Tool to download / merge individual RNASeq files from the GDC Portal into a matrices identified by TCGA barcode.
Inputs and Outputs:
| I/O | File |
|---|---|
| Input | GDC Manifest File |
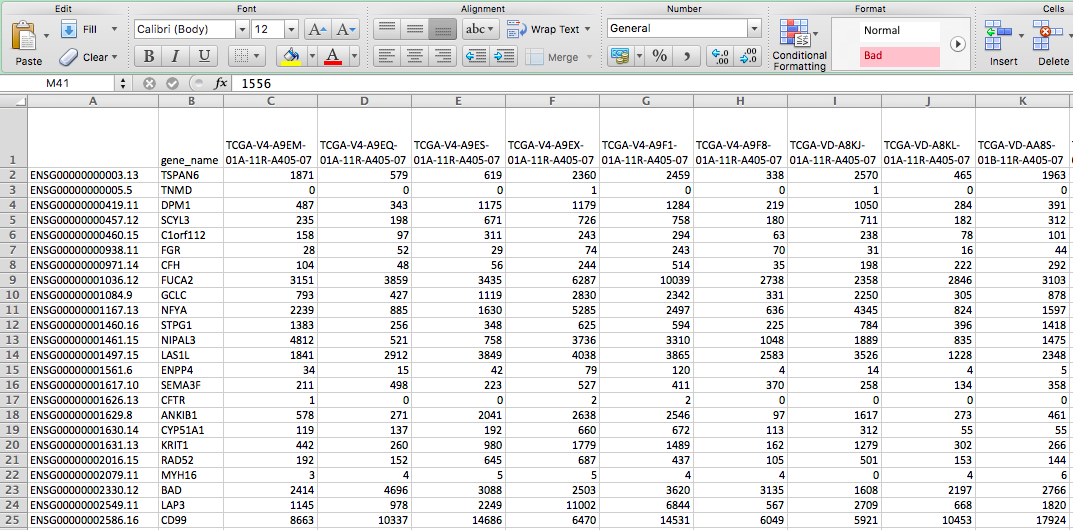
| Output | Merged_Counts.tsv (HTSeq - Counts) |
| Merged_FPKM.tsv (HTSeq - FPKM) | |
| Merged_FPKM-UQ.tsv (HTSeq - FPKM-UQ) | |
| Merged_miRNA_Counts.tsv | |
| Merged_miRNA_rpmm.tsv |
Bioinformatics Pipeline Information:
Requirements:
- Python 3+
- pandas ( https://pandas.pydata.org/pandas-docs/stable/install.html ):
pip3 install pandas
Quick Start:
- Download
gdc-rnaseq-tool.pypython script - Download manifest containing RNA/miRNA expression files from https://portal.gdc.cancer.gov/
python3 gdc-rnaseq-tool.py <manifest_file>
The GDC RNASeq tool produces matrices of merged RNA/MiRNA expression data given a manifest file.
Usage: python3 gdc-rnaseq-tool.py <manifest_file>
Notes:
- A test manifest is provided for troubleshooting:
python3 gdc-rnaseq-tool.py Test_Manifest.txt - Files are by default downloaded to the same folder as the manifest file that was provided
Release Notes:
Version 1.0: Feb 8, 2018
- Initial release
Known Issues: N/A