The goal of mcmodel is to standardize the method for melting curve
data modelling and prediction.
You can install the development version of mcmodel from GitHub with:
# install.packages("devtools")
devtools::install_github("gaospecial/mcmodel")library(mcmodel)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(readr)
library(ggplot2)# sample data
file = system.file("quantstudio", "quantstudio-export.txt.gz", package = 'mcmodel')
all_data = read_quantstudio(file)all_data contains all information in that file, including experimental
setting, melting curve data, and so on.
all_data
#> An object of class 'QuantStudioRaw':
#> Slots: [Sample Setup], [Raw Data], [Amplification Data], [Multicomponent Data], [Results], [Reagent Information], [Melt Curve Raw Data], Meta;We use get_quantstudio_melting_curve() to get melting curve dataset.
melting_curve_data = get_quantstudio_melting_curve(all_data)
melting_curve_data
#> # A tibble: 88,854 × 6
#> well well_position reading temperature fluorescence derivative
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 1 A1 1 75.1 1.60 0.043
#> 2 1 A1 2 75.1 1.60 0.037
#> 3 1 A1 3 75.2 1.60 0.032
#> 4 1 A1 4 75.3 1.61 0.029
#> 5 1 A1 5 75.4 1.61 0.027
#> 6 1 A1 6 75.5 1.60 0.028
#> 7 1 A1 7 75.5 1.60 0.031
#> 8 1 A1 8 75.6 1.59 0.036
#> 9 1 A1 9 75.7 1.59 0.041
#> 10 1 A1 10 75.8 1.59 0.046
#> # ℹ 88,844 more rowsIn this experiment, these are > 81k observations of six variables.
If you want to plot the melting curve, just use the
plot_quantstudio_melting_curve().
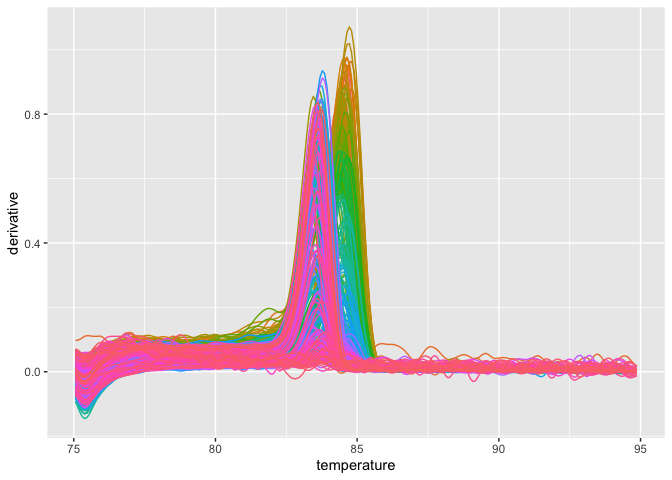
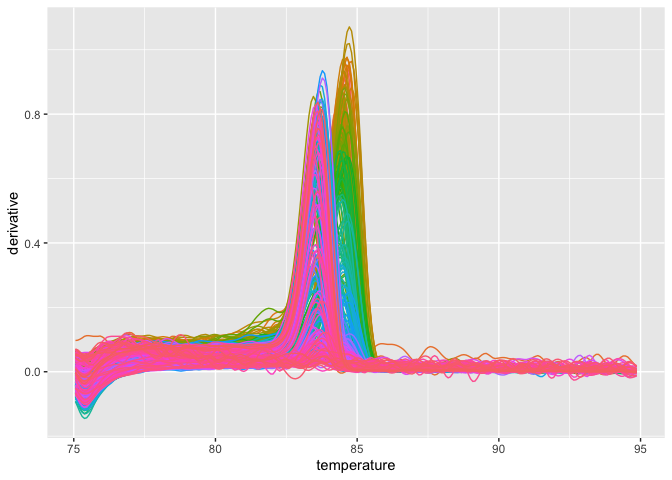
plot_quantstudio_melting_curve(melting_curve_data)Type ?plot_quantstudio_melting_curve in your console, and you may find
that there is a extra argument for this function.
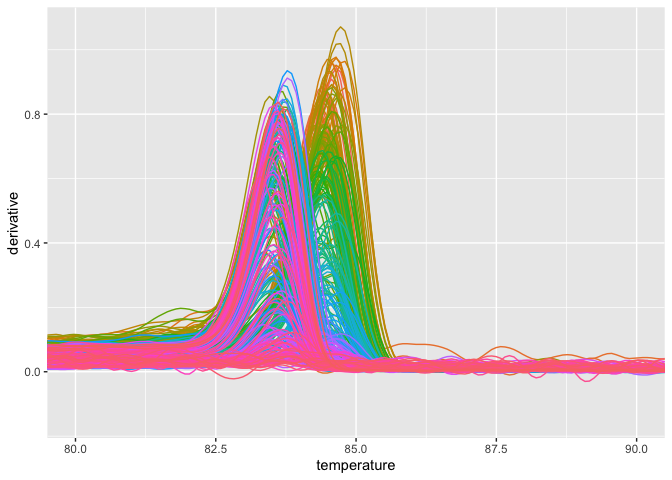
plot_quantstudio_melting_curve(melting_curve_data, y = "derivative")If you want to scale up this plot,
plot_quantstudio_melting_curve(melting_curve_data, y = "derivative") +
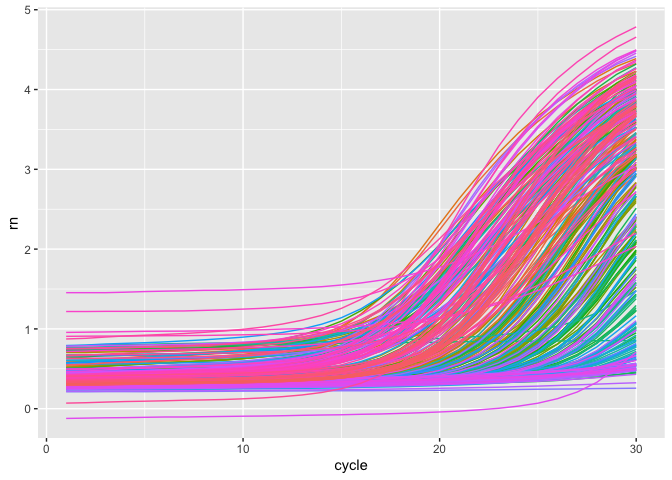
coord_cartesian(xlim = c(80, 90))Likewise, you may get the amplification data, and plot it.
amplification_data = get_quantstudio_amplication(all_data)
plot_quantstudio_amplification_curve(amplification_data)
#> Warning: Removed 900 rows containing missing values (`geom_line()`).This is just a small step to proceed this program, and it is the way what I want.
空白的平板,这样画:
plot_384() +
geom_point(color = "grey", size = 5)画单一物种的数量。首先画大肠杆菌的含量(log2)。
Plate.
file = system.file("quantstudio", "quantstudio-plate.csv.gz", package = 'mcmodel')
concentration = read_csv(file)
#> Rows: 352 Columns: 3
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (1): well_position
#> dbl (2): label_E, label_P
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
concentration
#> # A tibble: 352 × 3
#> well_position label_E label_P
#> <chr> <dbl> <dbl>
#> 1 A1 1 0
#> 2 A2 1 0
#> 3 A3 1 0
#> 4 A4 0 1
#> 5 A5 0 1
#> 6 A6 0 1
#> 7 A7 1 1
#> 8 A8 1 0.5
#> 9 A9 1 0.25
#> 10 A10 1 0.125
#> # ℹ 342 more rowsplot_384_single_concentration(concentration, species = "label_E")其次,用另外一个颜色画假单胞菌的含量。
plot_384_single_concentration(concentration, species = "label_P")画每个孔的结构。
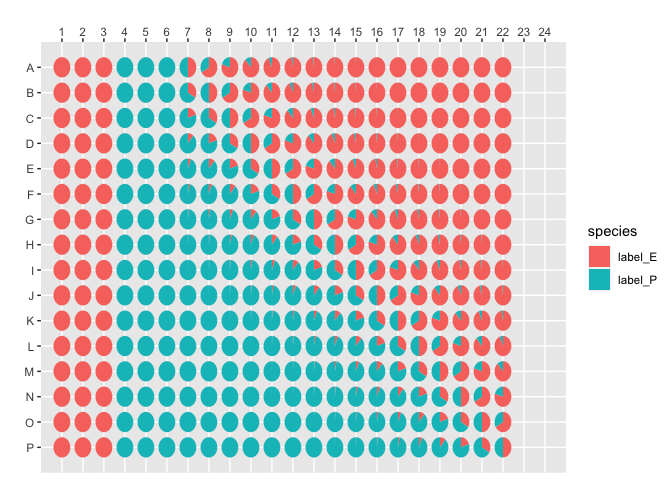
plot_384_community_structure(concentration)使用 MeltingCurve 对象可以确保数据不会被意外修改。
创建 MC 对象
从实验原始数据创建 MC 对象。
all_data
#> An object of class 'QuantStudioRaw':
#> Slots: [Sample Setup], [Raw Data], [Amplification Data], [Multicomponent Data], [Results], [Reagent Information], [Melt Curve Raw Data], Meta;
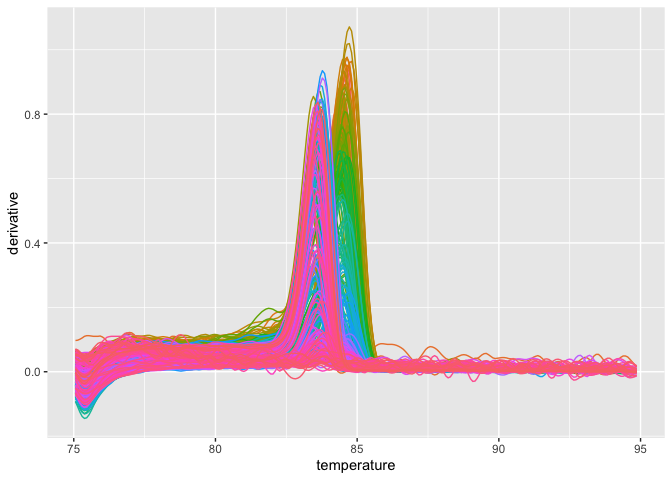
mc = quantstudio2mc(all_data)画熔解曲线。
plot_mc(mc, show_tm = FALSE)过滤熔解曲线的温度范围,以及反应孔。
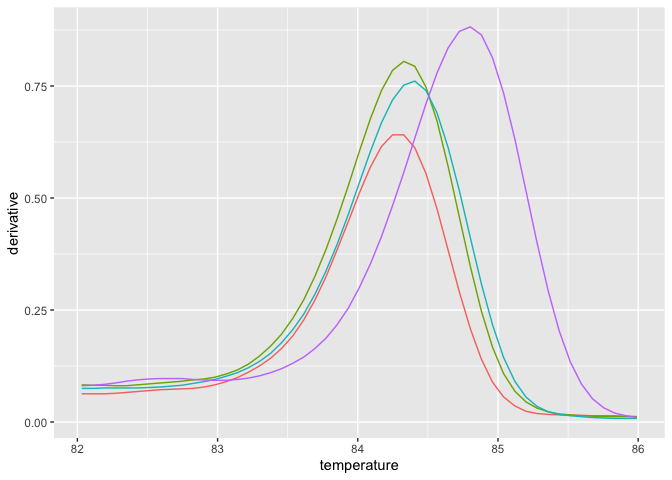
mc_narrow = filterData(mc, 82, 86, c("A1","A2","B1","B2"))
plot_mc(mc_narrow, show_tm = FALSE)对熔解曲线进行重采样。
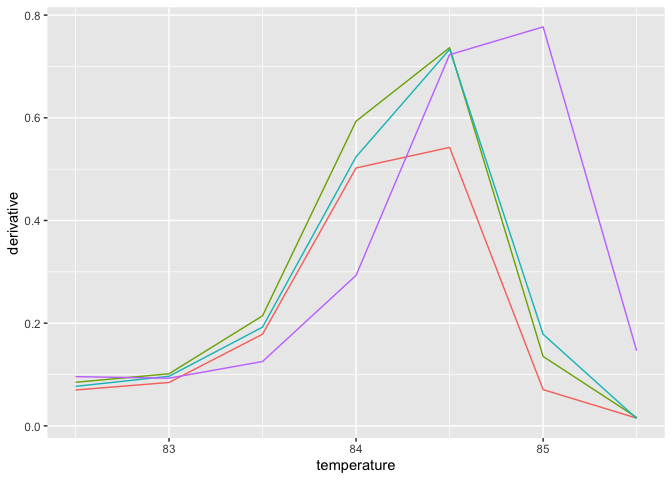
mc_low = transformData(mc_narrow, step = 0.5)
plot_mc(mc_low)将对象转变成建模数据集。
df = mc_tbl2wider(mc_low)
df
#> # A tibble: 4 × 8
#> well_position T82.5 T83 T83.5 T84 T84.5 T85 T85.5
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 A1 0.0698 0.0845 0.179 0.502 0.542 0.0707 0.0151
#> 2 A2 0.0849 0.101 0.214 0.593 0.737 0.135 0.0163
#> 3 B1 0.0769 0.0965 0.192 0.524 0.733 0.178 0.0145
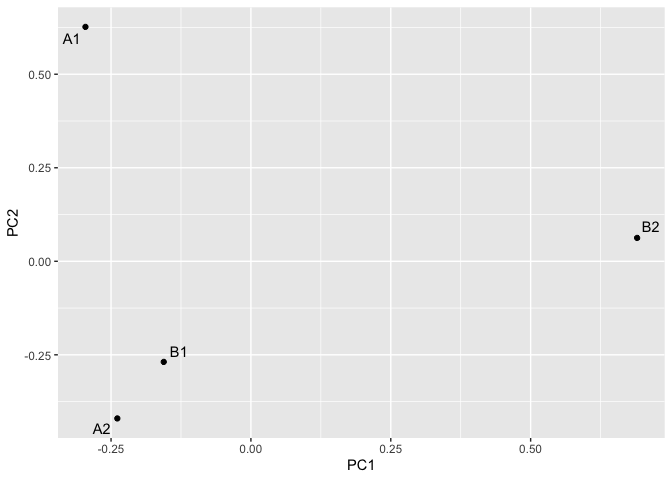
#> 4 B2 0.0959 0.0928 0.125 0.293 0.723 0.777 0.146主成分分析。
pca = mc_pca(mc_low)
mc_pca_plot(pca)
#> Warning: Duplicated aesthetics after name standardisation: NA