pyg_spectral is a PyTorch Geometric-based framework for analyzing, implementing, and benchmarking spectral GNNs with effectiveness and efficiency evaluations. Our preliminary paper is available on arXiv. Artifact and additional results can be found in the Appendix.
Important
Why this project?
We list the following highlights of our framework compared to PyG and similar works:
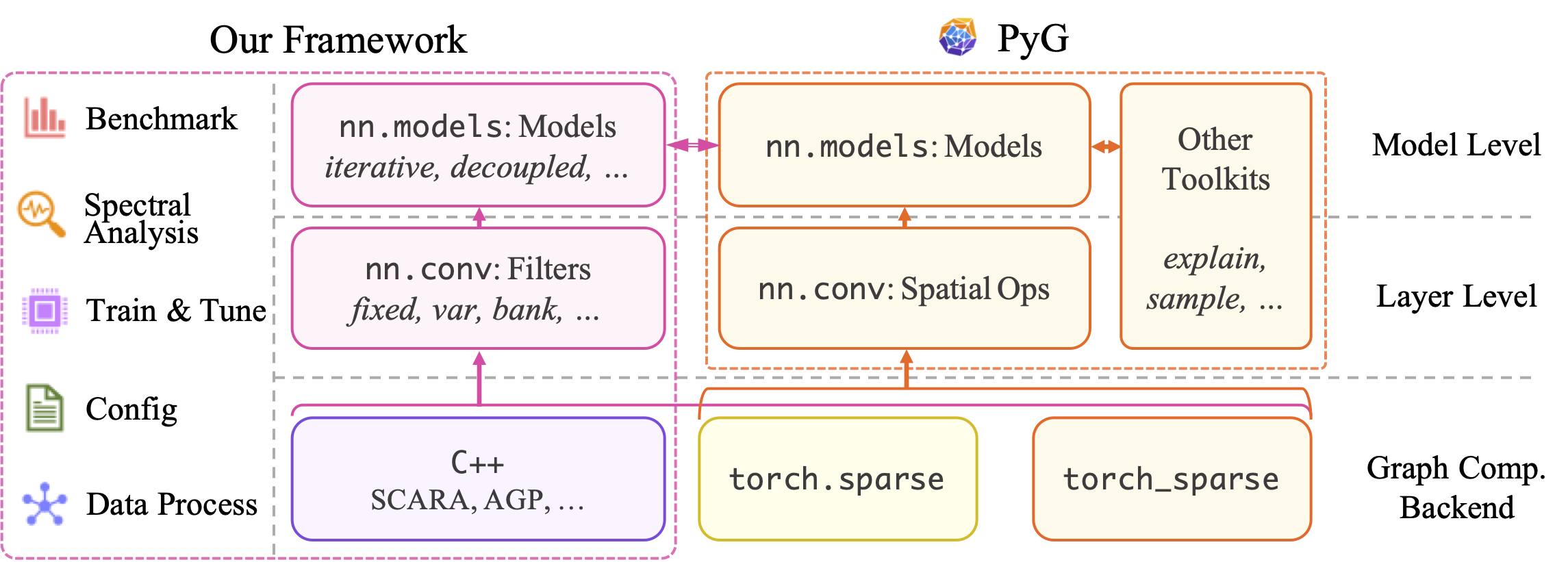
- Unified Framework: We offer a plug-and-play collection for spectral models and filters in unified and efficient implementations, rather than a model-specific design. Our rich collection greatly extends the PyG model zoo.
- Spectral-oriented Design: We decouple non-spectral designs and feature the pivotal spectral kernel being consistent throughout different settings. Most filters are thus easily adaptable to a wide range of model-level options, including those provided by PyG and PyG-based frameworks.
- High scalability: As spectral GNNs are inherently suitable for large-scale learning, our framework is feasible to common scalable learning schemes and acceleration techniques. Several spectral-oriented approximation algorithms are also supported.
This package can be easily installed by running pip at package root path:
pip install -r requirements.txt
pip install -e .[benchmark]The installation script already covers the following core dependencies:
- PyTorch (
>=2.01) - PyTorch Geometric (
>=2.5.3) - TorchMetrics (
>=1.0): only required forbenchmark/experiments. - Optuna (
>=3.4): only required for hyperparameter search inbenchmark/experiments.
For additional installation of the C++ backend, please refer to propagations/README.md.
Acquire results on the effectiveness and efficiency of spectral GNNs. Datasets will be automatically downloaded and processed by the code.
cd benchmark
bash scripts/runfb.shbash scripts/runmb.shbash scripts/eval_degng.shFigures can be plotted by: benchmark/notebook/fig_degng.ipynb.
bash scripts/eval_hop.shFigures can be plotted by: benchmark/notebook/fig_hop.ipynb.
bash scripts/exp_regression.shRefer to the help text by:
python benchmark/run_single.py --helpusage: python run_single.py
options:
--help show this help message and exit
# Logging configuration
--seed SEED random seed
--dev DEV GPU id
--suffix SUFFIX Result log file name. None:not saving results
-quiet File log. True:dry run without saving logs
--storage {state_file,state_ram,state_gpu}
Checkpoint log storage scheme.
--loglevel LOGLEVEL Console log. 10:progress, 15:train, 20:info, 25:result
# Data configuration
--data DATA Dataset name
--data_split DATA_SPLIT Index or percentage of dataset split
--normg NORMG Generalized graph norm
--normf [NORMF] Embedding norm dimension. 0: feat-wise, 1: node-wise, None: disable
# Model configuration
--model MODEL Model class name
--conv CONV Conv class name
--num_hops NUM_HOPS Number of conv hops
--in_layers IN_LAYERS Number of MLP layers before conv
--out_layers OUT_LAYERS Number of MLP layers after conv
--hidden_channels HIDDEN Number of hidden width
--dropout_lin DP_LIN Dropout rate for linear
--dropout_conv DP_CONV Dropout rate for conv
# Training configuration
--epoch EPOCH Number of epochs
--patience PATIENCE Patience epoch for early stopping
--period PERIOD Periodic saving epoch interval
--batch BATCH Batch size
--lr_lin LR_LIN Learning rate for linear
--lr_conv LR_CONV Learning rate for conv
--wd_lin WD_LIN Weight decay for linear
--wd_conv WD_CONV Weight decay for conv
# Model-specific
--theta_scheme THETA_SCHEME Filter name
--theta_param THETA_PARAM Hyperparameter for filter
--combine {sum,sum_weighted,cat}
How to combine different channels of convs
# Conv-specific
--alpha ALPHA Decay factor
--beta BETA Scaling factor
# Test flags
--test_deg Call TrnFullbatch.test_deg()
In benchmark/trainer/load_data.py, append the SingleGraphLoader._resolve_import() method to include new datasets under respective protocols.
New spectral filters to pyg_spectral/nn/conv/ can be easily implemented by only three steps, then enjoys a range of model architectures, analysis utilities, and training schemes.
The base class BaseMP provides essential methods for building spectral filters. We can define a new filter class SkipConv by inheriting from it:
from torch import Tensor
from pyg_spectral.nn.conv.base_mp import BaseMP
class SkipConv(BaseMP):
def __init__(self, num_hops, hop, cached, **kwargs):
kwargs['propagate_mat'] = 'A-I'
super(SkipConv, self).__init__(num_hops, hop, cached, **kwargs)The propagation matrix is specified by the propagate_mat argument as a string. Each matrix can be the normalized adjacency matrix (A) or the normalized Laplacian matrix (L), with optional diagonal scaling, where the scaling factor can either be a number or an attribute name of the class. Multiple propagation matrices can be combined by ,. Valid examples: A, L-2*I, L,A+I,L-alpha*I.
Similar to PyG modules, our spectral filter class takes the graph attribute x and edge index edge_index as input. The _get_convolute_mat() method prepares the representation matrices used in recurrent computation as a dictionary:
def _get_convolute_mat(self, x, edge_index):
return {'x': x, 'x_1': x}The above example overwrites the method for SkipConv, returning the input feature x and a placeholder x_1 for the representation in the previous hop.
The _forward() method implements recurrent computation of the filter. Its input/output is a dictionary combining the propagation matrices defined by propagate_mat and the representation matrices prepared by _get_convolute_mat().
def _forward(self, x, x_1, prop):
if self.hop == 0:
# No propagation for k=0
return {'x': x, 'x_1': x, 'prop': prop}
h = self.propagate(prop, x=x)
h = h + x_1
return {'x': h, 'x_1': x, 'prop': prop}Similar to PyG modules, the propagate() method conducts graph propagation by the given matrices. The above example corresponds to the graph propagation with a skip connection to the previous representation:
Now the SkipConv filter is properly defined. The following snippet use the DecoupledVar model composing 10 hops of SkipConv filters, which can be used as a normal PyTorch model:
from pyg_spectral.nn.models import DecoupledVar
model = DecoupledVar(conv='SkipConv', num_hops=10, in_channels=x.size(1), hidden_channels=x.size(1), out_channels=x.size(1))
out = model(x, edge_index)| Category | Model |
|---|---|
| Fixed Filter | GCN, SGC, gfNN, GZoom, S²GC, GLP, APPNP, GCNII, GDC, DGC, AGP, GRAND+ |
| Variable Filter | GIN, AKGNN, DAGNN, GPRGNN, ARMAGNN, ChebNet, ChebNetII, HornerGCN / ClenshawGCN, BernNet, LegendreNet, JacobiConv, FavardGNN / OptBasisGNN |
| Filter Bank | AdaGNN, FBGNN, ACMGNN, FAGCN, G²CN, GNN-LF/HF, FiGURe |
| Source | Graph |
|---|---|
| PyG | cora, citeseer, pubmed, flickr, actor |
| OGB | ogbn-arxiv, ogbn-mag, ogbn-products |
| LINKX | penn94, arxiv-year, genius, twitch-gamer, snap-patients, pokec, wiki |
| Yandex | chameleon, squirrel, roman-empire, minesweeper, amazon-ratings, questions, tolokers |
benchmark/: codes for benchmark experiments.pyg_spectral/: core codes for spectral GNNs designs, arranged in PyG structure.nn.conv: spectral spectral filters, similar totorch_geometric.nn.conv.nn.models: common neural network architectures, similar totorch_geometric.nn.models.nn.propagations: C++ backend for efficient propagation algorithms.
log/: experiment log files and parameter search results.data/: raw and processed datasets arranged following different protocols.
- Support C++ propagation backend with efficient algorithms.
- Unifews
- SGC
- GBP, AGP
- Support more transformation operations.
- Generalize ACMGNN
- Support iterative eigen-decomposition for full-spectrum spectral filters.
- Jacobi method
- Lanczos method
- This project is licensed under the MIT LICENSE.
- Please export CITATION by using "Cite this repository" in the right sidebar.
Footnotes
-
Please refer to the official guide if a specific CUDA version is required for PyTorch. ↩