By Jiaqi Gu, Zhengqi Gao, Chenghao Feng, Hanqing Zhu, Ray T. Chen, Duane S. Boning and David Z. Pan.
This repo is the official implementation of "NeurOLight: A Physics-Agnostic Neural Operator Enabling Parametric Photonic Device Simulation".
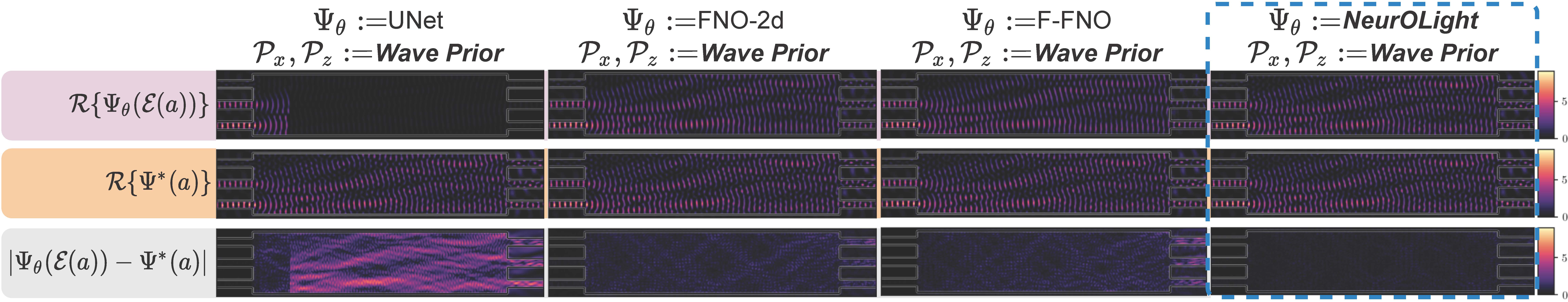
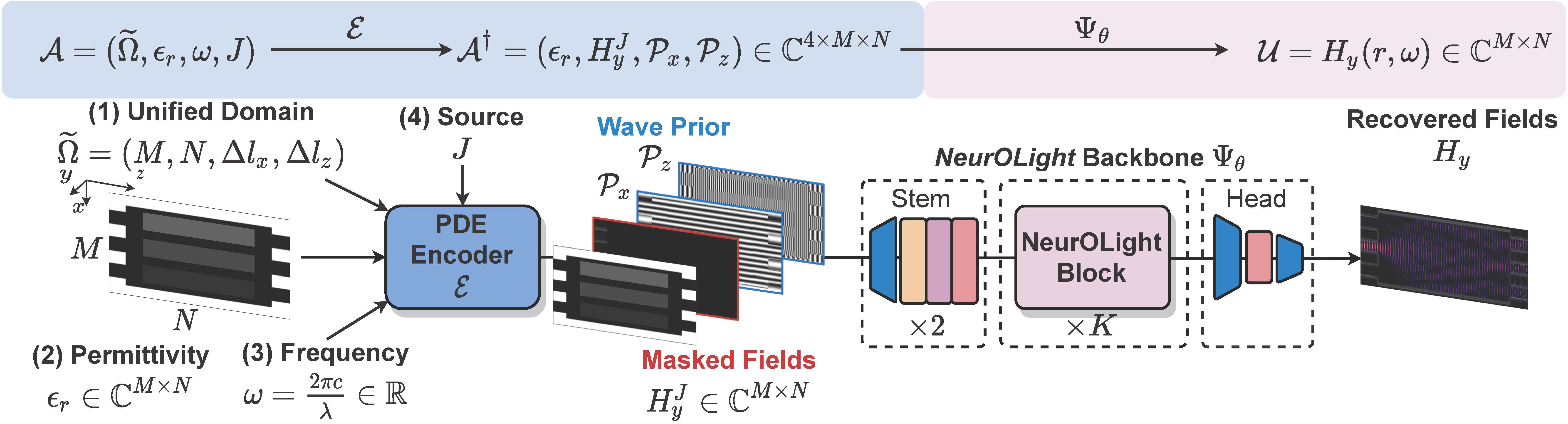
NeurOLight is a physics-agnostic neural operator-based framework to enable ultra-fast parametric integrated photonic device simulation.
NeurOLight learns a family of frequency-domain Maxwell PDEs to predict the light field insides integrated photonic devices.
We discretize different devices into a unified domain, represent parametric PDEs with a compact wave prior, and encode the incident light via masked source modeling.
NeurOLight is designed with parameter-efficient cross-shaped NeurOLight blocks and adopts superposition-based augmentation for data-efficient learning.
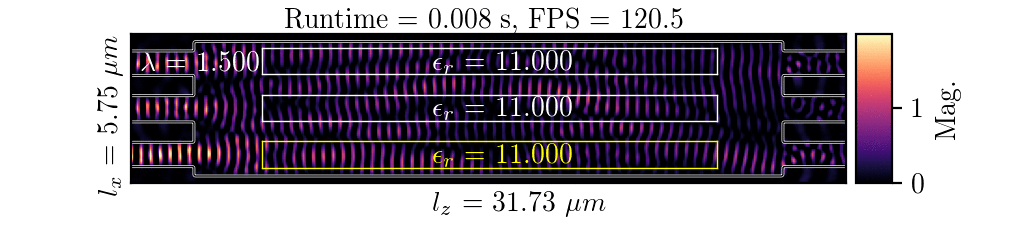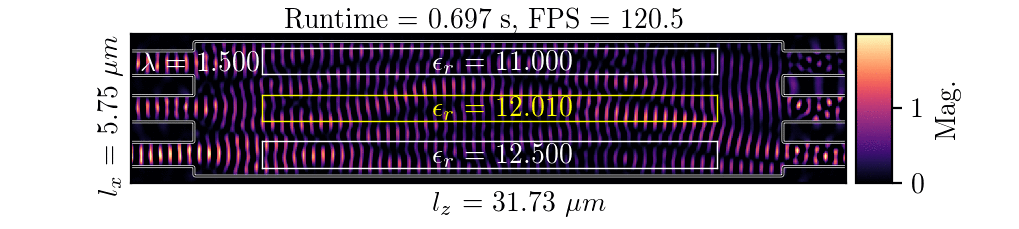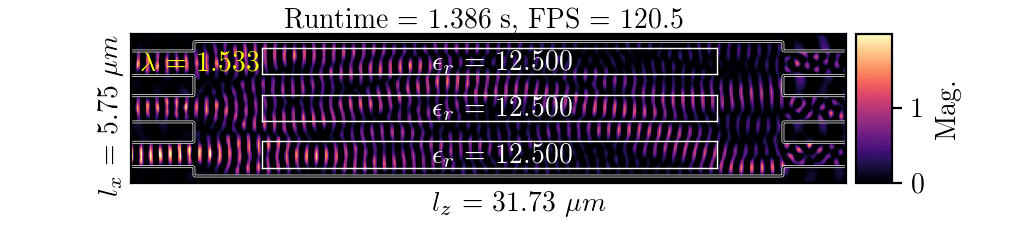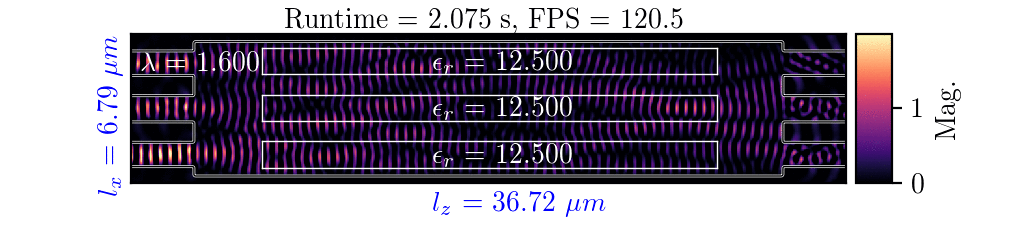
NeurOLight generalizes to a large space of unseen simulation settings, demonstrates 2-orders-of-magnitude faster simulation speed than numerical solvers, and outperforms prior neural network models by ~54% lower prediction error with ~44% fewer parameters.
- Python >= 3.6
- pyutils >= 0.0.1. See pyutils for installation.
- pytorch-onn >= 0.0.5. See pytorch-onn for installation.
- Python libraries listed in
requirements.txt - NVIDIA GPUs and CUDA >= 10.2
- configs/: configuration files
- core/
- models/
- layers/
- neurolight_conv2d: NeurOLight block definition
- fno_conv2d: FNO block definition
- factorfno_conv2d: Factorized-FNO block definition
- neurolight_cnn.py: NeurOLight model definition
- fno_cnn.py: FNO model definition
- factorfno_cnn.py: Factorized-FNO (F-FNO) model definition
- unet.py: UNet-2d model definition
- pde_base.py: base model definition
- utils.py: utility functions
- constant.py: constant definition
- layers/
- builder.py: build training utilities
- utils.py: customized loss function definition
- models/
- scripts/: contains experiment scripts
- data/: MMI simulation dataset
- train.py: training logic
- test.py: inference logic
- tune.py: transfer and finetune logic
-
3x3 tunable MMI dataset simulation and raw dataset generation (Optional).
DATASETneed to be replaced by one option from (mmi3x3,mmi4x4,mmi5x5,mmi3x3_etched)
data/mmi/> python3 make_dataset.py DATASET -
Train
MODELon 3x3 tunable MMI datasets, replaceMODELwith one option from (neurolight,fno,factorfno,unet),
> python3 scripts/mmi/MODEL/train_random_tunable.py -
Or, train
MODELon 3x3 etched MMI datasets, replaceMODELwith one option from (neurolight,fno,factorfno,unet),
> python3 scripts/mmi/MODEL/train_random_etched.py -
Test the trained model on test simulation instances with the single-shot multi-source mode. (Need to modify the path to trained checkpoint in the script).
python3 scripts/mmi/MODEL/test_main_results.py -
Checkpoints for 12-layer NeurOLight on tunable MMIs and 16-layer NeurOLight on etched MMIs are available at link
@inproceedings{gu2022NeurOLight,
title={NeurOLight: A Physics-Agnostic Neural Operator Enabling Parametric Photonic Device Simulation},
author={Jiaqi Gu and Zhengqi Gao and Chenghao Feng and Hanqing Zhu and Ray T. Chen and Duane S. Boning and David Z. Pan},
booktitle={Conference on Neural Information Processing Systems (NeurIPS)},
year={2022}
}