| output | |||
|---|---|---|---|
|
The gitlab version of the malariaAtlas package has some additional bugfixes over the stable CRAN package. If you have any issues, try installing the latest github version. See below for instructions.
This package allows you to download parasite rate data (Plasmodium falciparum and P. vivax), survey occurrence data of the 41 dominant malaria vector species, and modelled raster outputs from the Malaria Atlas Project.
More details and example analyses can be found in the [published paper)[(https://malariajournal.biomedcentral.com/articles/10.1186/s12936-018-2500-5).
The data can be explored at https://malariaatlas.org/explorer/#/explorer.
listData() retrieves a list of available data to download.
Use:
-
listData(datatype = "pr points") OR listPoints(sourcedata = "pr points") to see for which countries PR survey point data can be downloaded.
-
listData(datatype = "vector points") OR listPoints(sourcedata = "vector points") to see for which countries Vector survey data can be downloaded.
-
use listData(datatype = "raster") OR listRaster() to see rasters available to download.
-
use listData(datatype = "shape") OR listShp() to see shapefiles available to download.
listData(datatype = "pr points")listData(datatype = "vector points")listData(datatype = "raster")listData(datatype = "shape")isAvailable_pr confirms whether or not PR survey point data is available to download for a specified country.
Check whether PR data is available for Madagascar:
isAvailable_pr(country = "Madagascar")## Confirming availability of PR data for: Madagascar...
## PR points are available for Madagascar.
Check whether PR data is available for the United States of America
isAvailable_pr(ISO = "USA")## Confirming availability of PR data for: USA...
## Specified location not found, see below comments:
##
## Data not found for 'USA', did you mean UGA OR SAU?
isAvailable_vec confirms whether or not vector survey point data is available to download for a specified country.
Check whether vector data is available for Myanmar:
isAvailable_vec(country = "Myanmar")## Confirming availability of Vector data for: Myanmar...
## Vector points are available for Myanmar.
getPR() downloads all publicly available PR data points for a specified country and plasmodium species (Pf, Pv or BOTH) and returns this as a dataframe with the following format:
MDG_pr_data <- getPR(country = "Madagascar", species = "both")## Rows: 1,651
## Columns: 28
## $ dhs_id <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "…
## $ site_id <int> 6221, 6021, 15070, 15795, 7374, 13099, 9849, 21475, 15943, 7930, …
## $ site_name <chr> "Andranomasina", "Andasibe", "Ambohimarina", "Antohobe", "Ambohim…
## $ latitude <dbl> -18.7170, -19.8340, -18.7340, -19.7699, -25.0323, -18.7010, -18.7…
## $ longitude <dbl> 47.4660, 47.8500, 47.2520, 46.6870, 46.9960, 47.1660, 47.4905, 47…
## $ rural_urban <chr> "UNKNOWN", "UNKNOWN", "UNKNOWN", "UNKNOWN", "UNKNOWN", "UNKNOWN",…
## $ country <chr> "Madagascar", "Madagascar", "Madagascar", "Madagascar", "Madagasc…
## $ country_id <chr> "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "M…
## $ continent_id <chr> "Africa", "Africa", "Africa", "Africa", "Africa", "Africa", "Afri…
## $ month_start <int> 1, 3, 1, 7, 4, 1, 1, 7, 4, 7, 11, 4, 9, 7, 7, 3, 7, 3, 4, 6, 3, 7…
## $ year_start <int> 1987, 1987, 1987, 1995, 1986, 1987, 1987, 1995, 1986, 1995, 1997,…
## $ month_end <int> 1, 3, 1, 8, 6, 1, 1, 8, 4, 8, 11, 6, 9, 8, 8, 6, 7, 3, 6, 6, 6, 7…
## $ year_end <int> 1987, 1987, 1987, 1995, 1986, 1987, 1987, 1995, 1986, 1995, 1997,…
## $ lower_age <dbl> 0, 0, 0, 2, 7, 0, 0, 2, 6, 2, 2, 7, 0, 2, 2, 0, 0, 0, 7, 0, 0, 0,…
## $ upper_age <int> 99, 99, 99, 9, 22, 99, 99, 9, 12, 9, 9, 22, 99, 9, 9, 5, 99, 99, …
## $ examined <int> 50, 246, 50, 50, 119, 50, 50, 50, 20, 50, 61, 156, 104, 50, 50, 1…
## $ positive <dbl> 7.5, 126.0, 2.5, 6.0, 37.0, 13.5, 4.5, 11.5, 8.0, 7.0, 3.0, 97.0,…
## $ pr <dbl> 0.1500, 0.5122, 0.0500, 0.1200, 0.3109, 0.2700, 0.0900, 0.2300, 0…
## $ species <chr> "P. falciparum", "P. falciparum", "P. falciparum", "P. falciparum…
## $ method <chr> "Microscopy", "Microscopy", "Microscopy", "Microscopy", "Microsco…
## $ rdt_type <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "…
## $ pcr_type <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ malaria_metrics_available <chr> "true", "true", "true", "true", "true", "true", "true", "true", "…
## $ location_available <chr> "true", "true", "true", "true", "true", "true", "true", "true", "…
## $ permissions_info <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "…
## $ citation1 <chr> "Lepers, J.P., Ramanamirija, J.A., Andriamangatiana-Rason, M.D. a…
## $ citation2 <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "…
## $ citation3 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
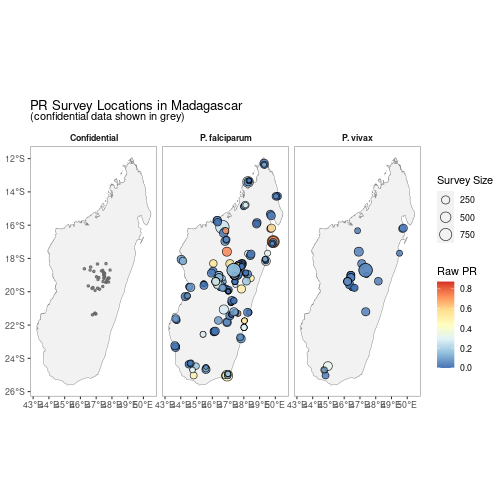
autoplot.pr.points configures autoplot method to enable quick mapping of the locations of downloaded PR points.
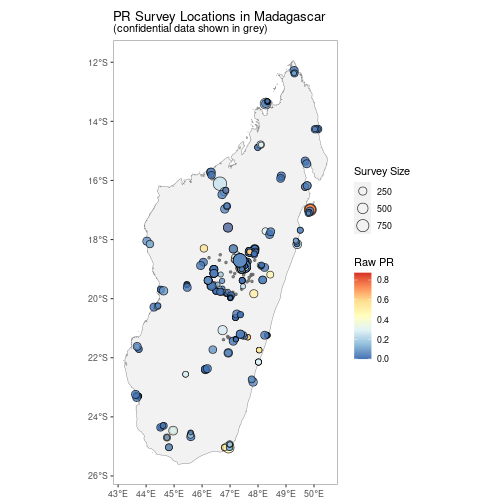
autoplot(MDG_pr_data)A version without facetting is also available.
autoplot(MDG_pr_data,
facet = FALSE)getVecOcc() downloads all publicly available Vector survey points for a specified country and and returns this as a dataframe with the following format:
MMR_vec_data <- getVecOcc(country = "Myanmar")## Rows: 2,866
## Columns: 24
## $ site_id <int> 30243, 30243, 30243, 30243, 1000000072, 1000000071, 1000000071, 1000000072, …
## $ latitude <dbl> 16.257, 16.257, 16.257, 16.257, 17.350, 17.380, 17.380, 17.350, 17.380, 17.3…
## $ longitude <dbl> 97.725, 97.725, 97.725, 97.725, 96.041, 96.037, 96.037, 96.041, 96.037, 96.0…
## $ country <chr> "Myanmar", "Myanmar", "Myanmar", "Myanmar", "Myanmar", "Myanmar", "Myanmar",…
## $ country_id <chr> "MMR", "MMR", "MMR", "MMR", "MMR", "MMR", "MMR", "MMR", "MMR", "MMR", "MMR",…
## $ continent_id <chr> "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asi…
## $ month_start <int> 2, 3, 8, 9, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 10, 5, 5, 5, 5, 5, 5, 5, …
## $ year_start <int> 1998, 1998, 1998, 1998, 1998, 1998, 1998, 1998, 1998, 1998, 1998, 1998, 1998…
## $ month_end <int> 2, 3, 8, 9, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 10, 3, 3, 3, 3, 3, 3, 3, …
## $ year_end <int> 1998, 1998, 1998, 1998, 2000, 2000, 2000, 2000, 2000, 2000, 2000, 2000, 2000…
## $ anopheline_id <int> 17, 17, 17, 17, 50, 49, 17, 51, 11, 4, 15, 1, 35, 30, 50, 51, 30, 17, 17, 11…
## $ species <chr> "Anopheles dirus species complex", "Anopheles dirus species complex", "Anoph…
## $ species_plain <chr> "Anopheles dirus", "Anopheles dirus", "Anopheles dirus", "Anopheles dirus", …
## $ id_method1 <chr> "unknown", "unknown", "unknown", "unknown", "morphology", "morphology", "mor…
## $ id_method2 <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", …
## $ sample_method1 <chr> "man biting", "man biting", "man biting", "man biting", "man biting indoors"…
## $ sample_method2 <chr> "animal baited net trap", "animal baited net trap", "animal baited net trap"…
## $ sample_method3 <chr> "", "", "", "", "animal baited net trap", "animal baited net trap", "animal …
## $ sample_method4 <chr> "", "", "", "", "house resting inside", "house resting inside", "house resti…
## $ assi <chr> "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", "", …
## $ citation <chr> "Oo, T.T., Storch, V. and Becker, N. (2003). <b><i>Anopheles</i> <i>dirus</…
## $ geom <chr> "POINT (16.257 97.725)", "POINT (16.257 97.725)", "POINT (16.257 97.725)", "…
## $ time_start <chr> "1998-02-01", "1998-03-01", "1998-08-01", "1998-09-01", "1998-05-01", "1998-…
## $ time_end <chr> "1998-02-01", "1998-03-01", "1998-08-01", "1998-09-01", "2000-03-01", "2000-…
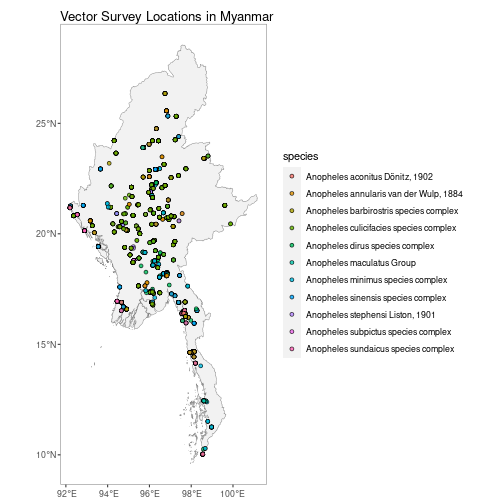
autoplot.vector.points configures autoplot method to enable quick mapping of the locations of downloaded vector points.
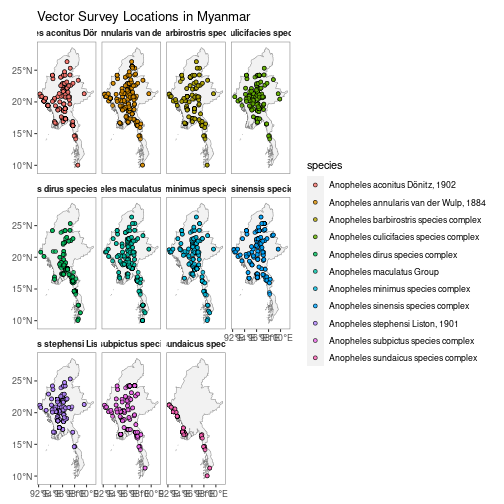
autoplot(MMR_vec_data)N.B. Facet-wrapped option is also available for species stratification.
autoplot(MMR_vec_data,
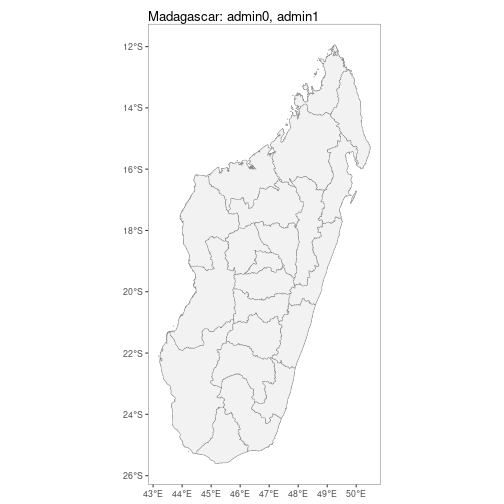
facet = TRUE)getShp() downloads a shapefile for a specified country (or countries) and returns this as either a spatialPolygon or data.frame object.
MDG_shp <- getShp(ISO = "MDG", admin_level = c("admin0", "admin1"))## Reading layer `mapadmin_0_2018' from data source
## `/tmp/Rtmpq4W3uO/shp/shp24fe071ca81346/mapadmin_0_2018.shp' using driver `ESRI Shapefile'
## Simple feature collection with 1 feature and 8 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: 43.19138 ymin: -25.60895 xmax: 50.48378 ymax: -11.94543
## Geodetic CRS: WGS 84
## Reading layer `mapadmin_1_2018' from data source
## `/tmp/Rtmpq4W3uO/shp/shp24fe071f231cf4/mapadmin_1_2018.shp' using driver `ESRI Shapefile'
## Simple feature collection with 22 features and 12 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: 43.19138 ymin: -25.60895 xmax: 50.48378 ymax: -11.94543
## Geodetic CRS: WGS 84
## Rows: 23
## Columns: 17
## $ iso <chr> "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", "MDG", …
## $ admn_level <dbl> 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1
## $ name_0 <chr> "Madagascar", "Madagascar", "Madagascar", "Madagascar", "Madagascar", "Madaga…
## $ id_0 <dbl> 10000910, 10000910, 10000910, 10000910, 10000910, 10000910, 10000910, 1000091…
## $ type_0 <chr> "Country", "Country", "Country", "Country", "Country", "Country", "Country", …
## $ name_1 <chr> "name_1", "Androy", "Anosy", "Atsimo Andrefana", "Atsimo Atsinanana", "Atsina…
## $ id_1 <chr> "id_1", "10023001", "10023002", "10023003", "10022990", "10023000", "10022994…
## $ type_1 <chr> "type_1", "Region", "Region", "Region", "Region", "Region", "Region", "Region…
## $ name_2 <chr> "name_2", "name_2", "name_2", "name_2", "name_2", "name_2", "name_2", "name_2…
## $ id_2 <chr> "id_2", "id_2", "id_2", "id_2", "id_2", "id_2", "id_2", "id_2", "id_2", "id_2…
## $ type_2 <chr> "type_2", "type_2", "type_2", "type_2", "type_2", "type_2", "type_2", "type_2…
## $ name_3 <chr> "name_3", "name_3", "name_3", "name_3", "name_3", "name_3", "name_3", "name_3…
## $ id_3 <chr> "id_3", "id_3", "id_3", "id_3", "id_3", "id_3", "id_3", "id_3", "id_3", "id_3…
## $ type_3 <chr> "type_3", "type_3", "type_3", "type_3", "type_3", "type_3", "type_3", "type_3…
## $ source <chr> "Madagascar NMCP 2016", "Madagascar NMCP 2016", "Madagascar NMCP 2016", "Mada…
## $ geometry <MULTIPOLYGON [°]> MULTIPOLYGON (((44.22776 -2..., MULTIPOLYGON (((45.25095 -2..., MULTIPOLYGON …
## $ country_level <chr> "MDG_0", "MDG_1", "MDG_1", "MDG_1", "MDG_1", "MDG_1", "MDG_1", "…
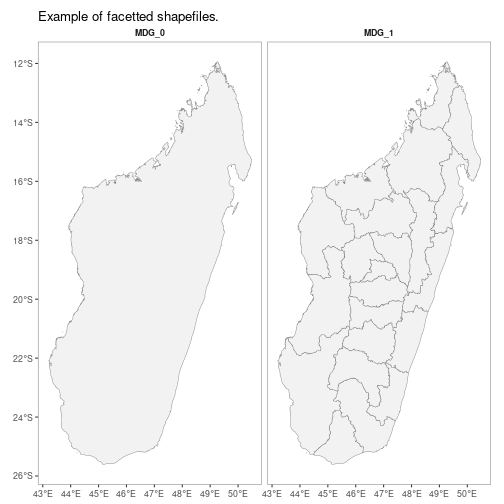
autoplot.sf configures autoplot method to enable quick mapping of downloaded shapefiles.
autoplot(MDG_shp)N.B. Facet-wrapped option is also available for species stratification.
autoplot(MDG_shp,
facet = TRUE,
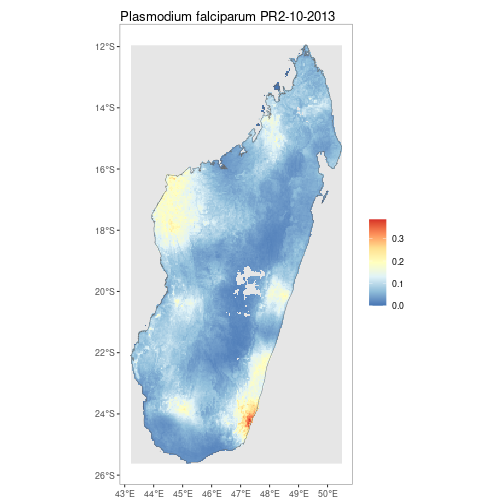
map_title = "Example of facetted shapefiles.")getRaster()downloads publicly available MAP rasters for a specific surface & year, clipped to a given bounding box or shapefile
MDG_shp <- getShp(ISO = "MDG", admin_level = "admin0")
MDG_PfPR2_10 <- getRaster(surface = "Plasmodium falciparum PR2-10", shp = MDG_shp, year = 2013)autoplot.SpatRaster & autoplot.SpatRasterCollection configures autoplot method to enable quick mapping of downloaded rasters.
p <- autoplot(MDG_PfPR2_10, shp_df = MDG_shp)By using the above tools along with ggplot, simple comparison figures can be easily produced.
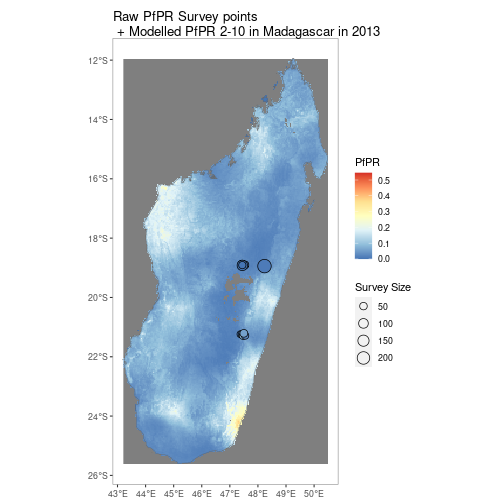
MDG_shp <- getShp(ISO = "MDG", admin_level = "admin0")
MDG_PfPR2_10 <- getRaster(surface = "Plasmodium falciparum PR2-10", shp = MDG_shp, year = 2013)
p <- autoplot(MDG_PfPR2_10, shp_df = MDG_shp, printed = FALSE)
pr <- getPR(country = c("Madagascar"), species = "Pf")
p[[1]] +
geom_point(data = pr[pr$year_start==2013,], aes(longitude, latitude, fill = positive / examined, size = examined), shape = 21)+
scale_size_continuous(name = "Survey Size")+
scale_fill_distiller(name = "PfPR", palette = "RdYlBu")+
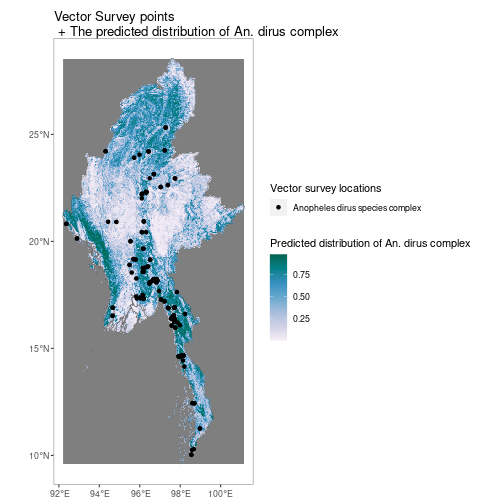
ggtitle("Raw PfPR Survey points\n + Modelled PfPR 2-10 in Madagascar in 2013")Similarly for vector survey data
MMR_shp <- getShp(ISO = "MMR", admin_level = "admin0")
MMR_An_dirus <- getRaster(surface = "Anopheles dirus species complex", shp = MMR_shp)
p <- autoplot(MMR_An_dirus, shp_df = MMR_shp, printed = FALSE)
vec <- getVecOcc(country = c("Myanmar"), species = "Anopheles dirus")
p[[1]] +
geom_point(data = vec, aes(longitude, latitude, colour = species))+
scale_colour_manual(values = "black", name = "Vector survey locations")+
scale_fill_distiller(name = "Predicted distribution of An. dirus complex", palette = "PuBuGn", direction = 1)+
ggtitle("Vector Survey points\n + The predicted distribution of An. dirus complex")Just install using install.packages("malariaAtlas") or using the package manager in RStudio.
While this version is not as well-tested, it may include additional bugfixes not in the stable CRAN version. Install the devtools package and then install using devtools::install_github('malaria-atlas-project/malariaAtlas')
- Removed
rgdal,sp,rasterpackages dependencies and addedterra,sf,tidyterra,lifecycle - As a result
getShpwill now returnsfobjects - Similarly,
getRasterreturnsSpatRasterorSpatRasterCollection - Some functions have been deprecated. They will still run in this version, with a warning. Please remove them from your code, they will be completely removed in the future.
- Deprecated
as.MAPraster,as.MAPshp. They are no longer required for working withautoplot - Deprecated
autoplot_MAPraster. Useautoplotdirectly with the result ofgetRaster
- Deprecated