dxCompiler takes a pipeline written in the Workflow Description Language (WDL) or Common Workflow Language (CWL) and compiles it to an equivalent workflow on the DNAnexus platform. The following standards are supported:
- WDL: draft-2, 1.0, and 1.1
- CWL: 1.2
Support for WDL 2.0 (aka 'development') is under active development and not yet production-ready. CWL 1.0 and 1.1 are not supported but can be upgraded to 1.2 to be compiled (see Preprocess CWL workflow).
To compile and run your workflow, make sure you have the following ready:
- DNAnexus platform account
- dx-toolkit
- Log in using
dx login - It is recommended to pre-select the project where you want to your compiled workflows to go using
dx select
- Log in using
- Java 8 or 11
- The latest dxCompiler JAR file from the releases page.
- docker, if you want to invoke dxCompiler with the run-dxcompiler-docker script using a public
dnanexus/dxcompilerdocker container. - Python 3.x to run the dxCompiler integration tests
To compile CWL tools/workflows, you might also need:
- sbpack to pack the workflow made up of multiple files into a single compound JSON document before compilation
- cwl-utils which includes a collection of Python scripts for loading and parsing CWL files
- cwl-upgrader to upgrade your workflow to version 1.2
- cwltool which provides comprehensive validation of CWL files as well as other tools related to working with CWL
dxCompiler uses wdlTools, a parser that adheres strictly to the WDL specifications. Most of the problematic automatic type conversions that are allowed by some other WDL runtime engines are not allowed by dxCompiler. Please use the command line tools in wdlTools (e.g. check and lint) to validate your WDL files before trying to compile them with dxCompiler.
The bam_chrom_counter workflow is written in WDL. Task slice_bam splits a bam file into an array of sub-files. Task
count_bam counts the number of alignments on a bam file. The workflow takes an input bam file, calls slice_bam to split it into chromosomes, and calls count_bam in parallel on each chromosome. The results comprise a bam index file, and an array with the number of reads per chromosome.
version 1.0
workflow bam_chrom_counter {
input {
File bam
}
call slice_bam {
input : bam = bam
}
scatter (slice in slice_bam.slices) {
call count_bam {
input: bam = slice
}
}
output {
File bai = slice_bam.bai
Array[Int] count = count_bam.count
}
}
task slice_bam {
input {
File bam
Int num_chrom = 22
}
command <<<
set -ex
samtools index ~{bam}
mkdir slices/
for i in `seq ~{num_chrom}`; do
samtools view -b ~{bam} -o slices/$i.bam $i
done
>>>
runtime {
docker: "quay.io/biocontainers/samtools:1.12--hd5e65b6_0"
}
output {
File bai = "~{bam}.bai"
Array[File] slices = glob("slices/*.bam")
}
}
task count_bam {
input {
File bam
}
command <<<
samtools view -c ~{bam}
>>>
runtime {
docker: "quay.io/biocontainers/samtools:1.12--hd5e65b6_0"
}
output {
Int count = read_int(stdout())
}
}From the command line, we can compile the workflow to the DNAnexus platform using the dxCompiler jar file.
$ java -jar dxCompiler.jar compile bam_chrom_counter.wdl -project project-xxxx -folder /my/workflows/
This compiles the source WDL file to several platform objects in the specified DNAnexus project project-xxxx under folder /my/workflows/
- A workflow
bam_chrom_counter - Two applets that can be called independently:
slice_bam, andcount_bam - A few auxiliary applets that process workflow inputs, outputs, and launch the scatter.
The generated workflow can be executed from the web UI (see instructions here) or via the DNAnexus command-line client. For example, to run the workflow with the input bam file project-BQbJpBj0bvygyQxgQ1800Jkk:file-FpQKQk00FgkGV3Vb3jJ8xqGV, use the following command:
dx run bam_chrom_counter -istage-common.bam=project-BQbJpBj0bvygyQxgQ1800Jkk:file-FpQKQk00FgkGV3Vb3jJ8xqGV
Alternatively, you can also convert a Cromwell JSON format input file into a DNAnexus format when compiling the workflow. Then you can pass the DNAnexus input file to dx run using -f option as described in detail here.
$ java -jar dxCompiler.jar compile bam_chrom_counter.wdl -project project-xxxx -folder /my/workflows/ -inputs bam_chrom_counter_input.json
$ dx run bam_chrom_counter -f bam_chrom_counter_input.dx.json
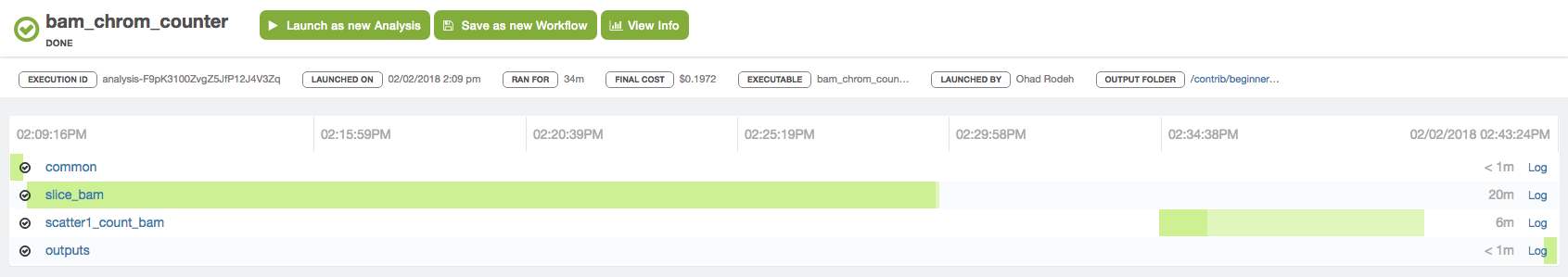
After launching the workflow analysis, you can monitor it on the CLI following these instructions or from the UI as suggested here. The snapshot below shows what you will see from the UI when the workflow execution is completed:
dxCompiler requires the source CWL file to be "packed" as a cwl.json file, which contains a single compound workflow with all the dependent processes included. Additionally, you may need to upgrade the version of your workflow to CWL v1.2.
We'll use the bam_chrom_counter CWL workflow similar to the WDL example above to illustrate upgrading, packing and running a CWL workflow. This workflow is written in CWL v1.0 and the top-level Workflow in bam_chrom_counter.cwl calls the two CommandLineTools in slice_bam.cwl and count_bam.cwl.
cwlVersion: v1.0
id: bam_chrom_counter
class: Workflow
requirements:
- class: ScatterFeatureRequirement
inputs:
- id: bam
type: File
# upload this local file to the platform and replace the path below with the DNAnexus URI "dx://project-xxx:file-yyyy"
default: "path/to/my/input_bam"
outputs:
- id: bai
type: File
outputSource: slice_bam/bai
- id: count
type: int[]
outputSource: count_bam/count
steps:
- id: slice_bam
run: slice_bam.cwl
in:
bam: bam
out: [bai, slices]
- id: count_bam
run: count_bam.cwl
scatter: bam
in:
bam: slice_bam/slices
out: [count]cwlVersion: v1.0
id: slice_bam
class: CommandLineTool
inputs:
- id: bam
type: File
- id: num_chrom
default: 22
type: int
outputs:
- id: bai
type: File
outputBinding:
glob: $(inputs.bam.basename).bai
- id: slices
type: File[]
outputBinding:
glob: "slices/*.bam"
requirements:
- class: InlineJavascriptRequirement
- class: ShellCommandRequirement
- class: DockerRequirement
dockerPull: "quay.io/biocontainers/samtools:1.12--hd5e65b6_0"
- class: InitialWorkDirRequirement
listing:
- entryname: slice_bam.sh
entry: |-
set -ex
samtools index $1
mkdir slices/
for i in `seq $2`; do
samtools view -b $1 -o slices/$i.bam $i
done
- entry: $(inputs.bam)
baseCommand: ["sh", "slice_bam.sh"]
arguments:
- position: 0
valueFrom: $(inputs.bam.basename)
- position: 1
valueFrom: $(inputs.num_chrom)
hints:
- class: NetworkAccess
networkAccess: true
- class: LoadListingRequirement
loadListing: deep_listingcwlVersion: v1.0
id: count_bam
class: CommandLineTool
requirements:
- class: InlineJavascriptRequirement
- class: ShellCommandRequirement
- class: DockerRequirement
dockerPull: "quay.io/biocontainers/samtools:1.12--hd5e65b6_0"
inputs:
- id: bam
type: File
inputBinding:
position: 1
baseCommand: ["samtools", "view", "-c"]
outputs:
- id: count
type: int
outputBinding:
glob: stdout
loadContents: true
outputEval: "$(parseInt(self[0].contents))"
stdout: stdout
hints:
- class: NetworkAccess
networkAccess: true
- class: LoadListingRequirement
loadListing: deep_listingBefore compilation, follow the steps below to preprocess these CWL files:
-
De-localize all local paths referenced in the CWL: if the CWL specifies a local path, e.g. a schema or a default value for a
file-type input (like the default path "path/to/my/input_bam" for inputbaminbam_chrom_counter.cwl), you need to upload this file to a DNAnexus project and then replace the local path in the CWL with its full DNAnexus URI, e.g.dx://project-XXX:file-YYY. -
Install
cwl-upgraderand upgrade the CWL files to v1.2 (needed in this case as CWL files are in CWL v1.0):$ pip3 install cwl-upgrader # upgrade all dependent CWL files, which will be saved in the current working directory $ cd contrib/beginner_example $ cwl-upgrader cwl_v1.0/bam_chrom_counter.cwl cwl_v1.0/slice_bam.cwl cwl_v1.0/count_bam.cwl -
Install
sbpackpackage and run thecwlpackcommand on the top-level workflow file to build a single packedbam_chrom_counter.cwl.jsonfile containing the top level workflow and all the steps it depends on:$ pip3 install sbpack $ cwlpack --add-ids --json bam_chrom_counter.cwl > bam_chrom_counter.cwl.json
dxCompiler compiles tools/workflows written according to the CWL v1.2 standard. You can use cwltool --validate to validate the packed CWL file you want to compile.
Once it is upgraded and packed as suggested above, we can compile it as a DNAnexus workflow and run it.
$ java -jar dxCompiler.jar compile bam_chrom_counter.cwl.json -project project-xxxx -folder /my/workflows/
$ dx run bam_chrom_counter -istage-common.bam=project-BQbJpBj0bvygyQxgQ1800Jkk:file-FpQKQk00FgkGV3Vb3jJ8xqGV
- WDL and CWL
- Calls with missing arguments have limited support
- All task and workflow names must be unique across the entire import tree
- For example, if
A.wdlimportsB.wdlandA.wdldefines workflowfoo, thenB.wdlcannot have a workflow or task namedfoo
- For example, if
- Subworkflows built from higher-level workflows are not intented to be used on their own
- WDL only
- Workflows with forward references (i.e. a variable referenced before it is declared) are not yet supported
- The alternative workflow output syntax that has been deprecated since WDL draft2 is not supported
- The
call ... aftersyntax introduced in WDL 1.1 is not yet supported
- CWL only
- Calling native DNAnexus apps/applets in CWL workflow using
dxniis not supported. SoftwareRequirementandInplaceUpdateRequirementare not yet supported- Publishing a dxCompiler-generated workflow as a global workflow is not supported
- Calling native DNAnexus apps/applets in CWL workflow using
- Advanced options explains additional compiler options
- Internals describes current compiler structure (work in progress)
- Tips examples for how to write good WDL code
- Debugging recommendations how to debug the workflows on DNAnexus platform
- A high-level list of changes between WDL draft-2 and version 1.0
See the development guide for more information on how to set up your development environment to contribute to dxCompiler and how to test your updates.
This software is a community effort! You can browse any of the contributions, that are not a part of dxCompiler main source codebase, below in our contrib folder, and add your own (see Contributing to dxCompiler).
Let us know if you would like to contribute, request a feature, or report a bug.