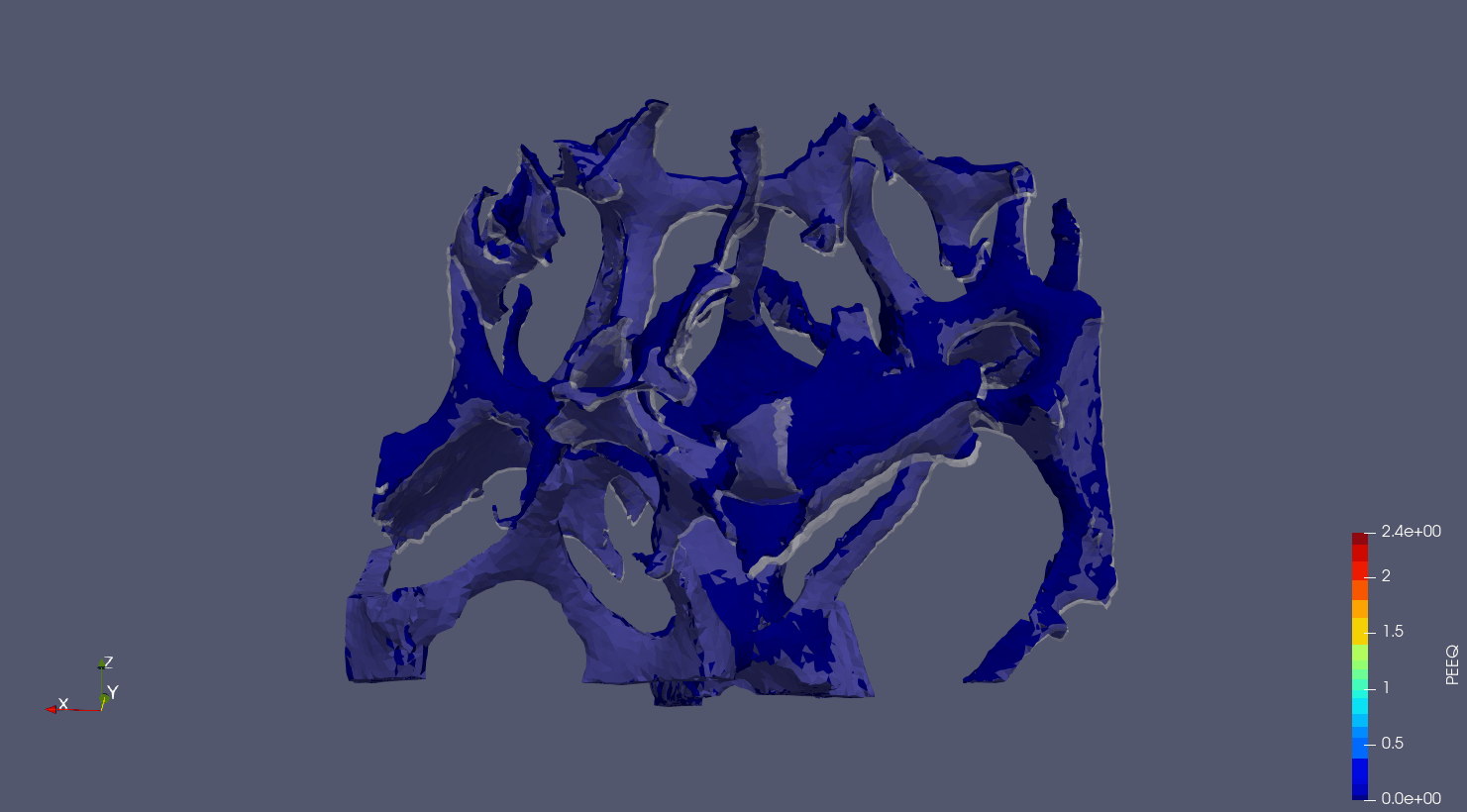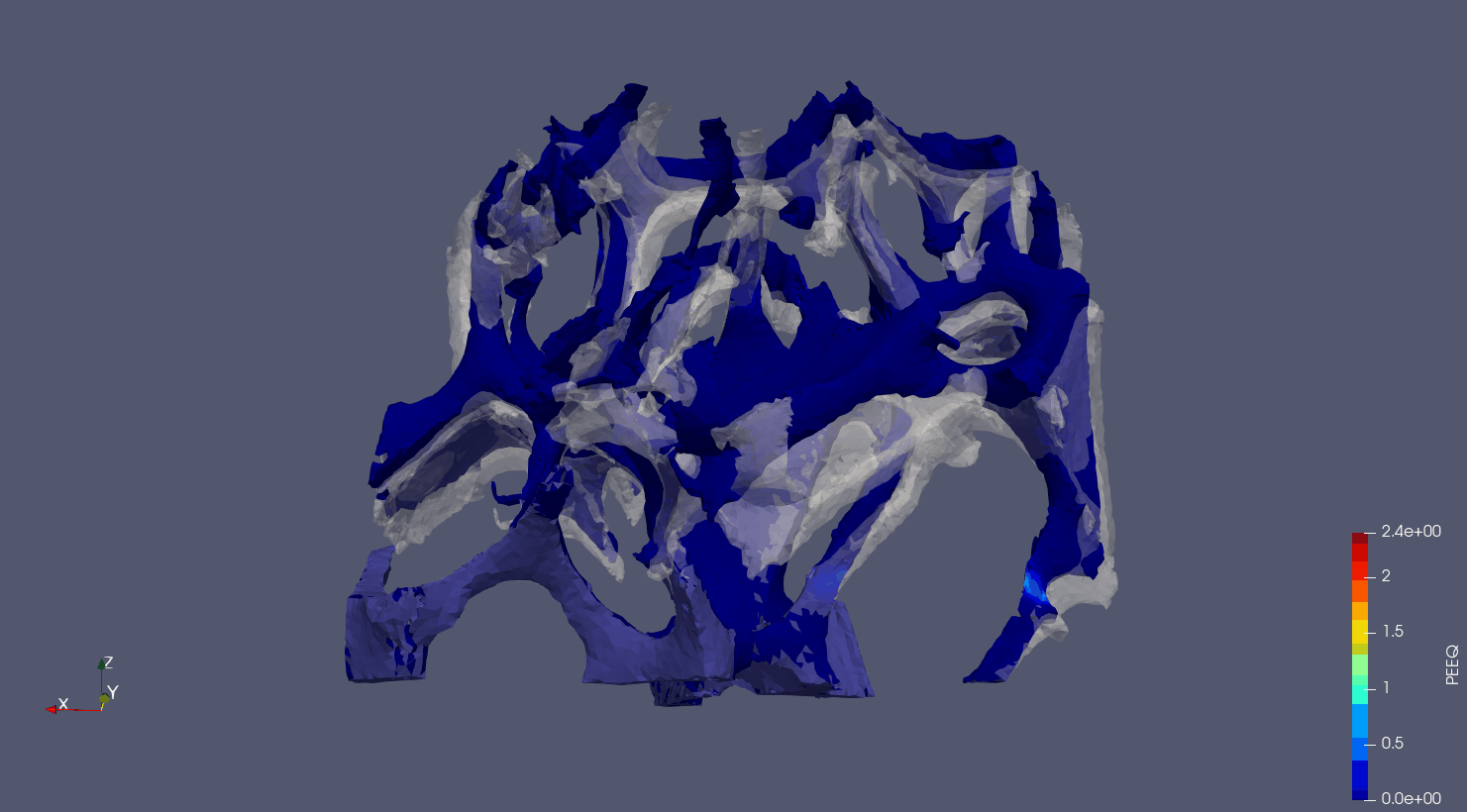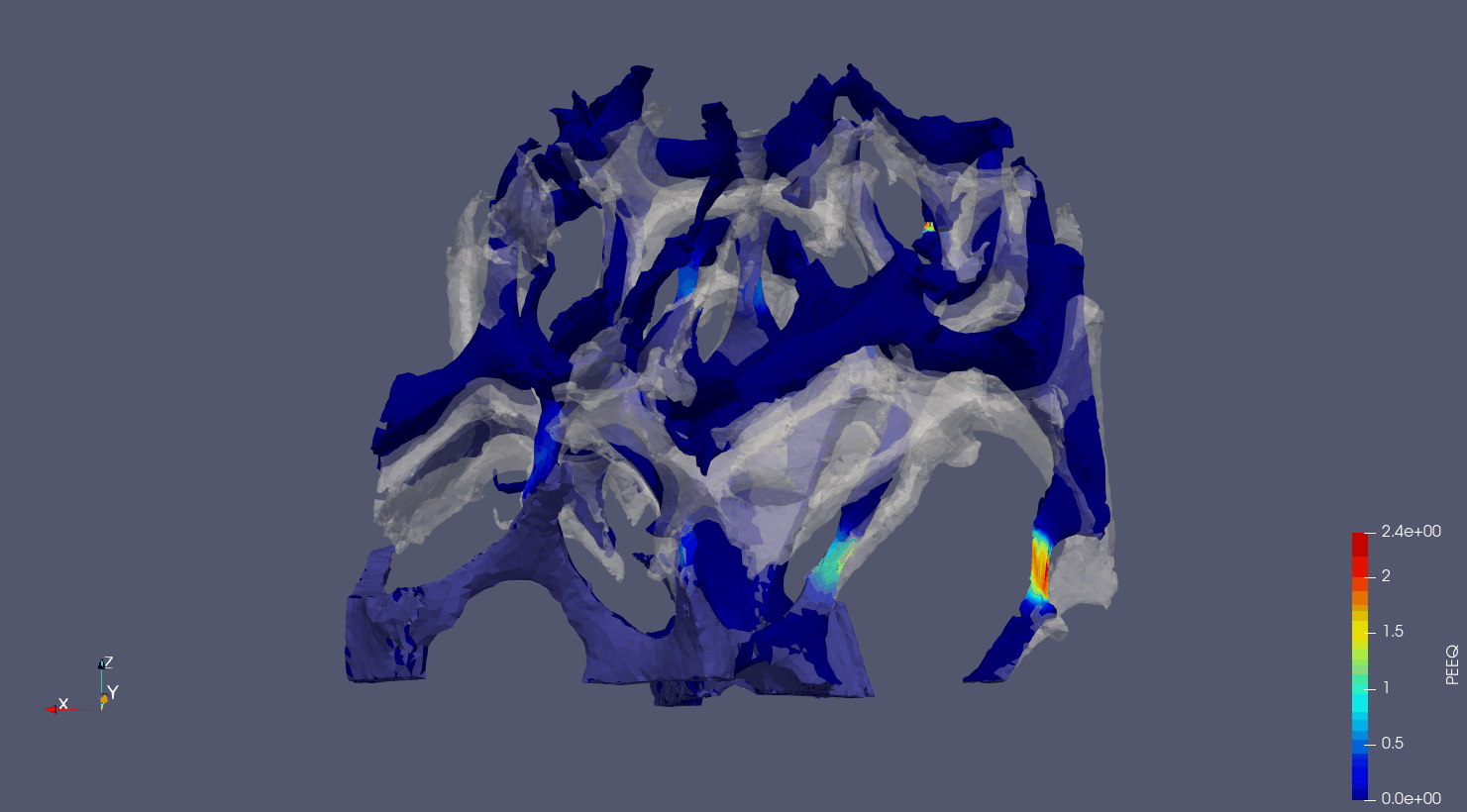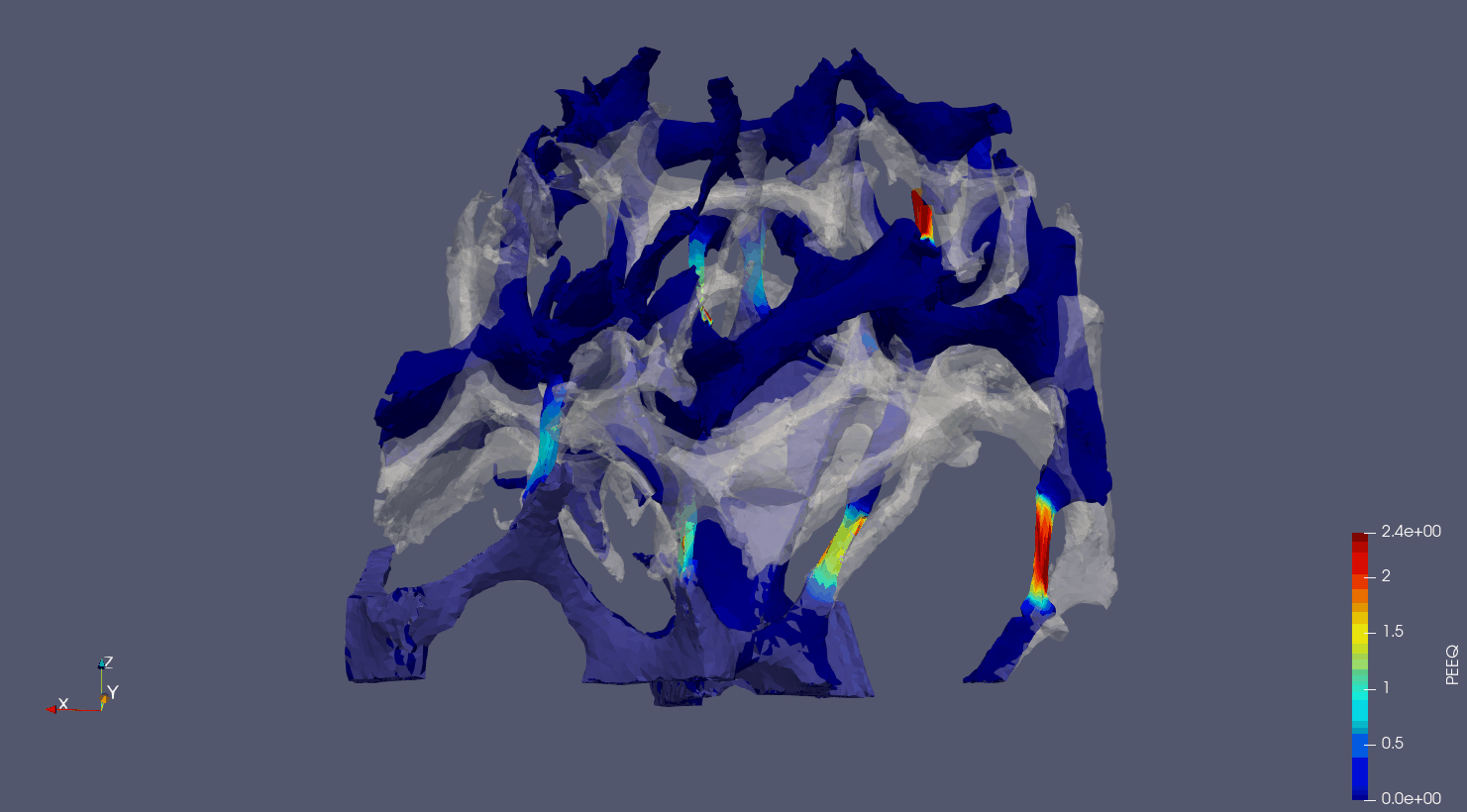Computed Tomography to Finite Elements.
Micro Finite Element (microFE) models can be derived from micro Computed Tomography (microCT) 3D images to non-destructively assess mechanical properties of biological or artificial specimens.
ciclope provides fully open-source pipelines from microCT data preprocessing to microFE model generation, solution and postprocessing.
For mesh generation, ciclope requires pygalmesh, a Python frontend to CGAL.
Follow the installation procedure for CGAL and Eigen.
After that, install pygalmesh with pip or conda
conda install -c conda-forge pygalmesh
After installing pygalmesh, you can install ciclope using pip. The flag [all] will install optional dependencies needed to run full pipelines and examples.
pip install ciclope[all]
Some examples will require DXchange. You can install it with
conda install -c conda-forge dxchangeTo verify your installation checkout this repository and run the tests with the command
cd test
python -m unittest -v test_ciclope.run_tests
If you want to contribute to this project, please install ciclope following the development guide.
ciclope pipelines can be run from the command line as a script. Scroll down and take a look at the Examples folder for this type of use. To view the command line script help run
ciclope -h
To use ciclope within python, import the package with
import ciclopeciclope.utils contains functions that help you read and pre-process 3D datasets for FE model generation.
Read 3D CT dataset stored as stack of TIFFs
from ciclope.utils.recon_utils import read_tiff_stack
input_file = './test_data/LHDL/3155_D_4_bc/cropped/3155_D_4_bc_0000.tif'
data_3D = read_tiff_stack(input_file)
vs = np.ones(3) * 0.06 # voxelsize [mm]read_tiff_stack reads all TIFF files (slices) contained in the input_file folder. The volume is stored in a 3D numpy.ndarray with size [slices, rows, columns].
Segment and remove unconnected voxels
from skimage import morphology
from ciclope.utils.preprocess import remove_unconnected
BW = data_3D > 142 # fixed global threshold
BW = morphology.closing(BW, morphology.ball(2)) # optional step
L = remove_unconnected(BW)If you already have a mesh file, you can skip the mesh generation steps and use ciclope with 3D meshio objects.
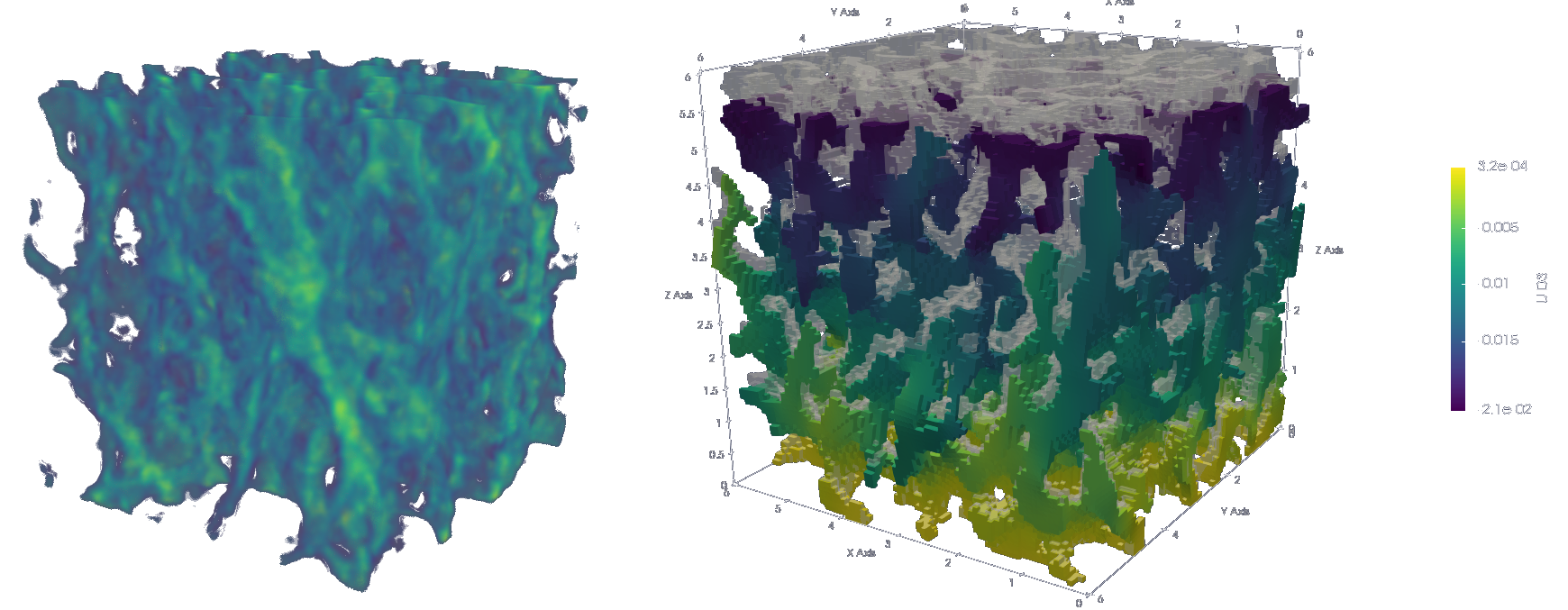 Generate unstructured grid mesh of hexahedra (voxels)
Generate unstructured grid mesh of hexahedra (voxels)
import ciclope
mesh = ciclope.core.voxelFE.vol2ugrid(data_3D, vs)Generate CalculiX input file .INP for voxel-FE model of linear elastic compression test
input_template = "./input_templates/tmp_example01_comp_static_bone.inp"
ciclope.core.voxelFE.mesh2voxelfe(mesh, input_template, 'foo.inp', keywords=['NSET', 'ELSET'])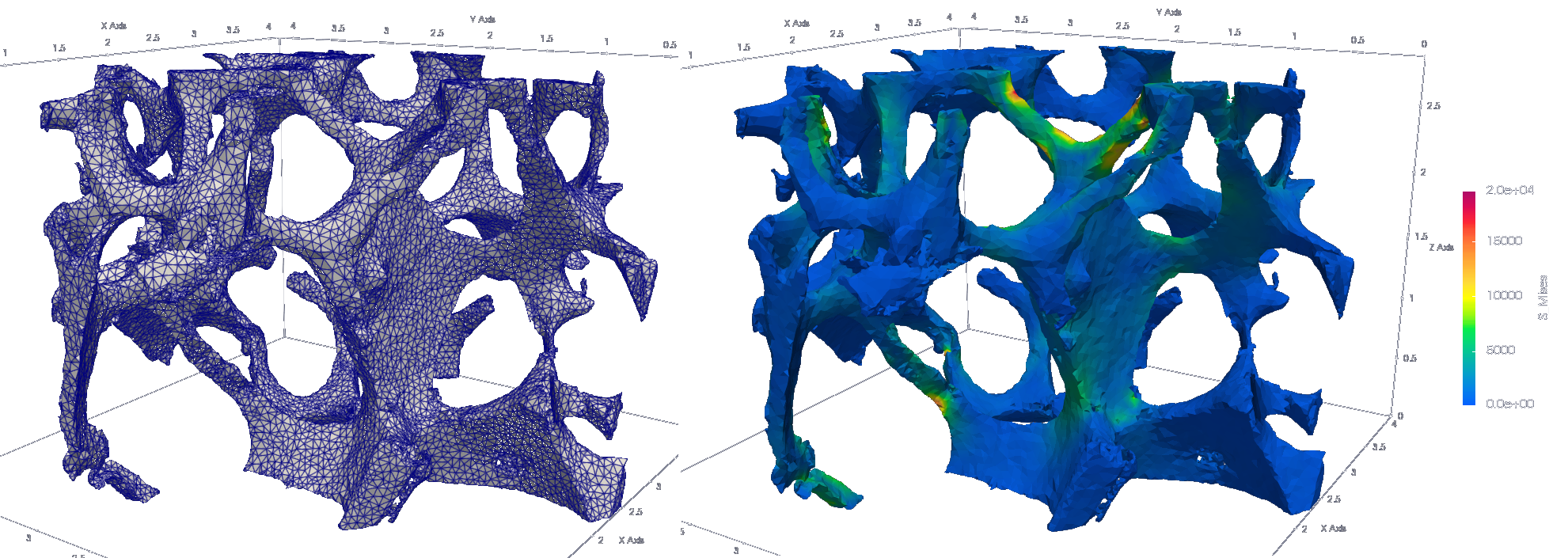 Generate mesh of tetrahedra.
Generate mesh of tetrahedra. ciclope uses pygalmesh for tetrahedra mesh generation
mesh = ciclope.core.tetraFE.cgal_mesh(L, vs, 'tetra', max_facet_distance=0.2, max_cell_circumradius=0.1)Generate CalculiX input file .INP for tetrahedra-FE model of non-linear tensile test
input_template = "./input_templates/tmp_example02_tens_static_steel.inp"
ciclope.core.tetraFE.mesh2tetrafe(mesh, input_template, 'foo.inp', keywords=['NSET', 'ELSET'])ciclope.utils.postprocess.paraviewplot calls ParaView to generate and save plots of a chosen model scalar field.
- Add path to your ParaView installation with
import sys
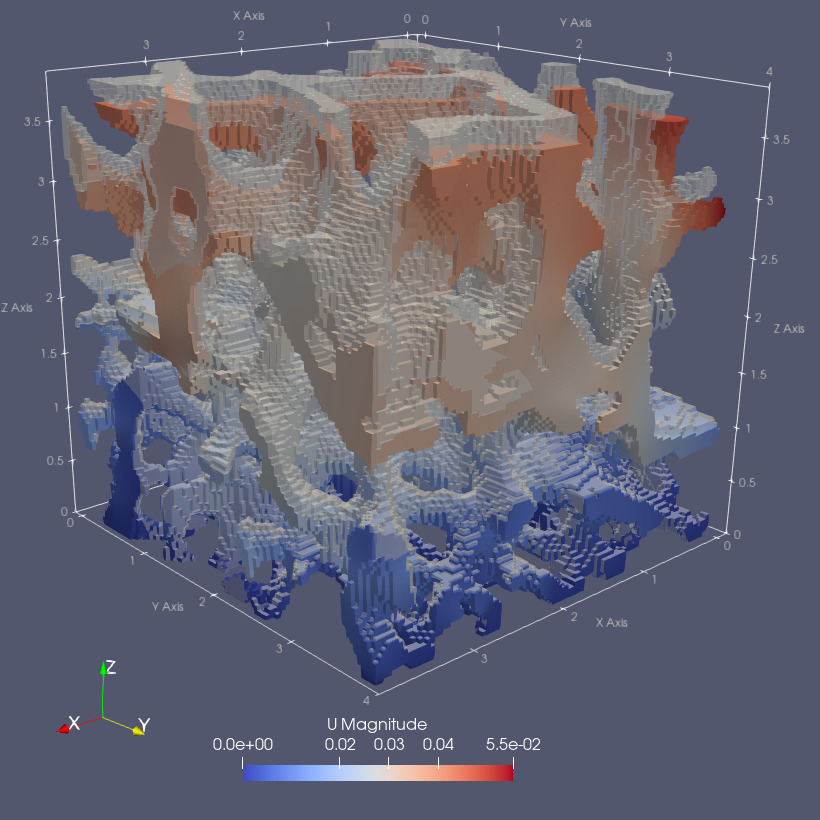
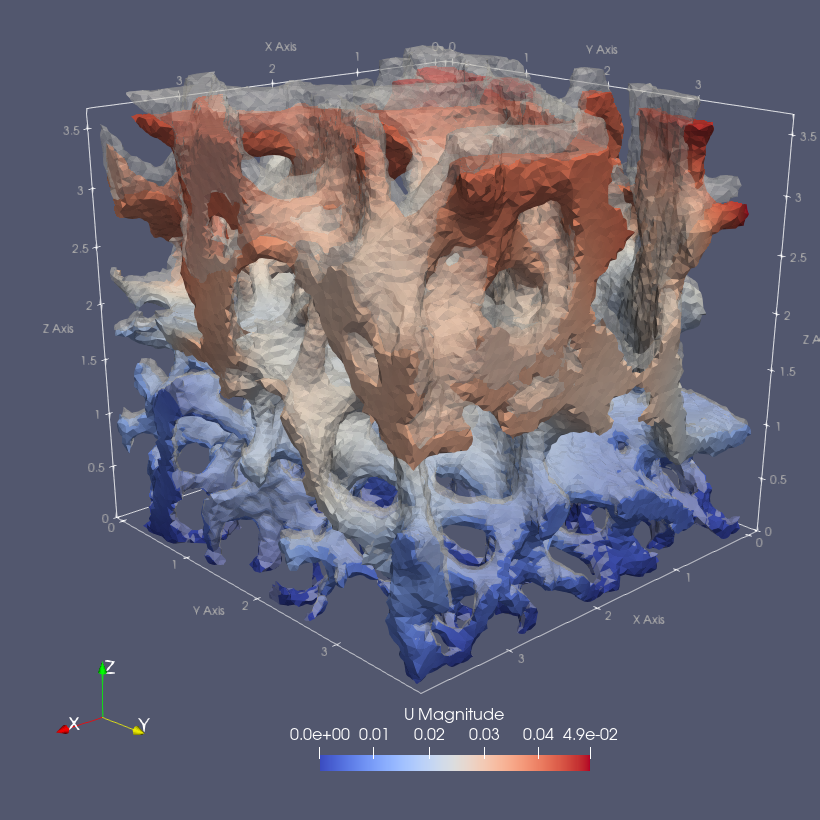
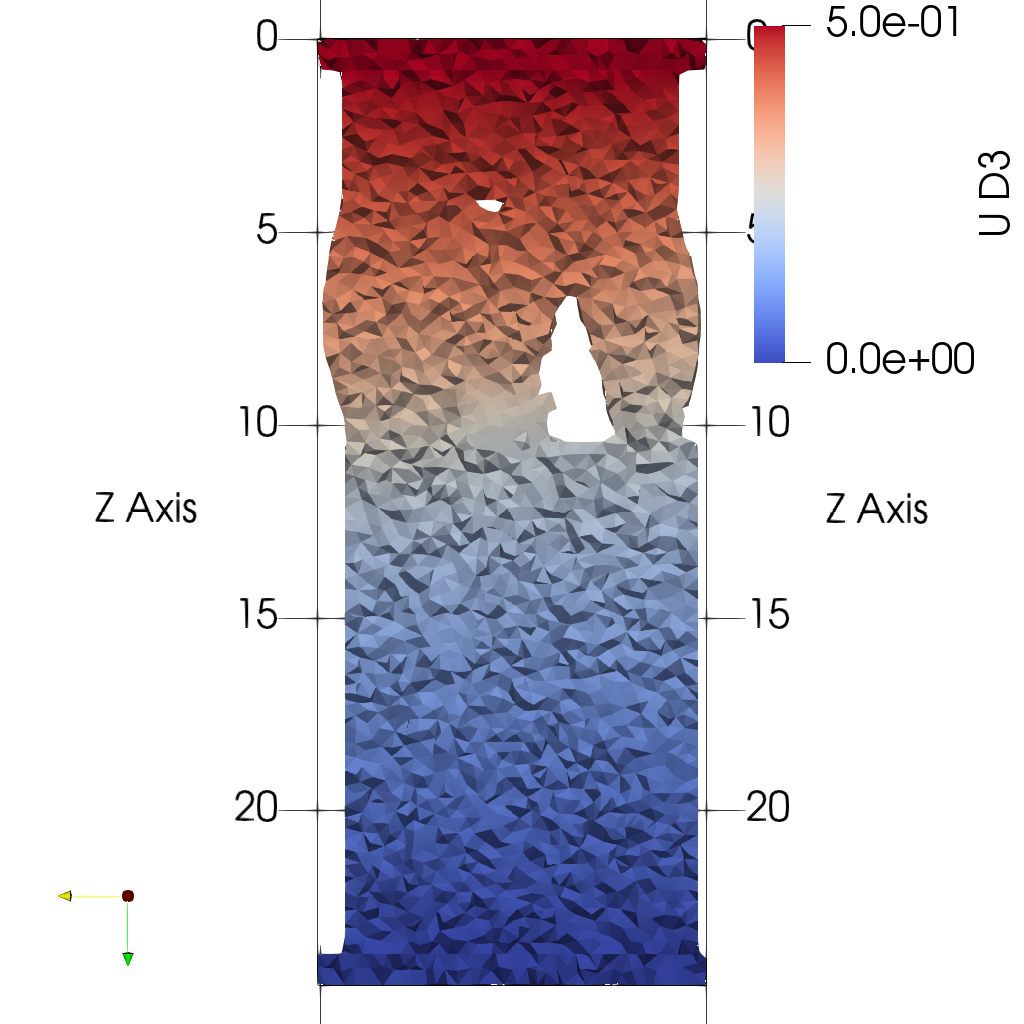
sys.path.append('~/Applications/ParaView-5.9.0-RC1-MPI-Linux-Python3.8-64bit/lib/python3.8/site-packages')- Plot midplanes of the vertical displacement field
UD3
ciclope.utils.postprocess.paraview_plot('test_data/tooth/results/Tooth_3_scaled_2.vtk', slicenormal="xyz",
RepresentationType="Surface", Crinkle=True, ColorBy=['U', 'D2'], Roll=90,
ImageResolution=[1024, 1024], TransparentBackground=True,
colormap='Cool to Warm')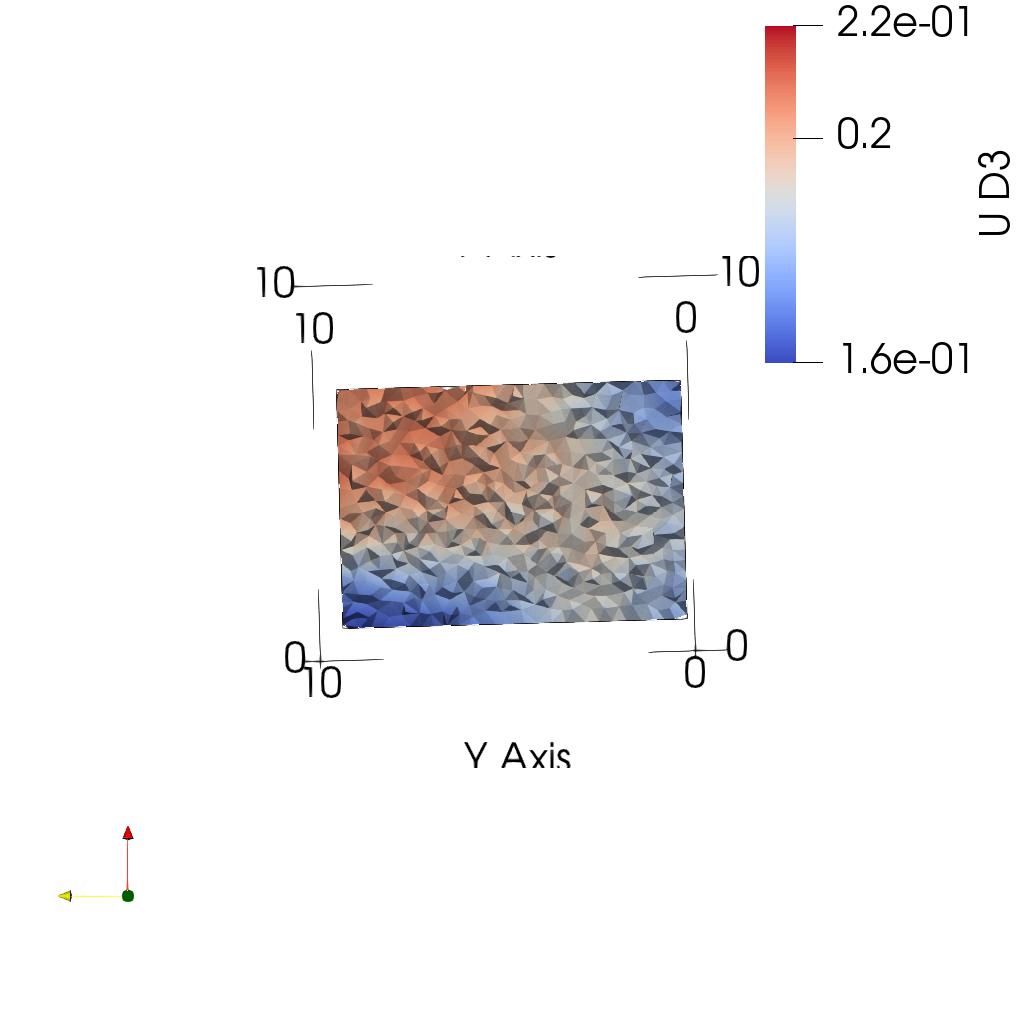 |
 |
 |
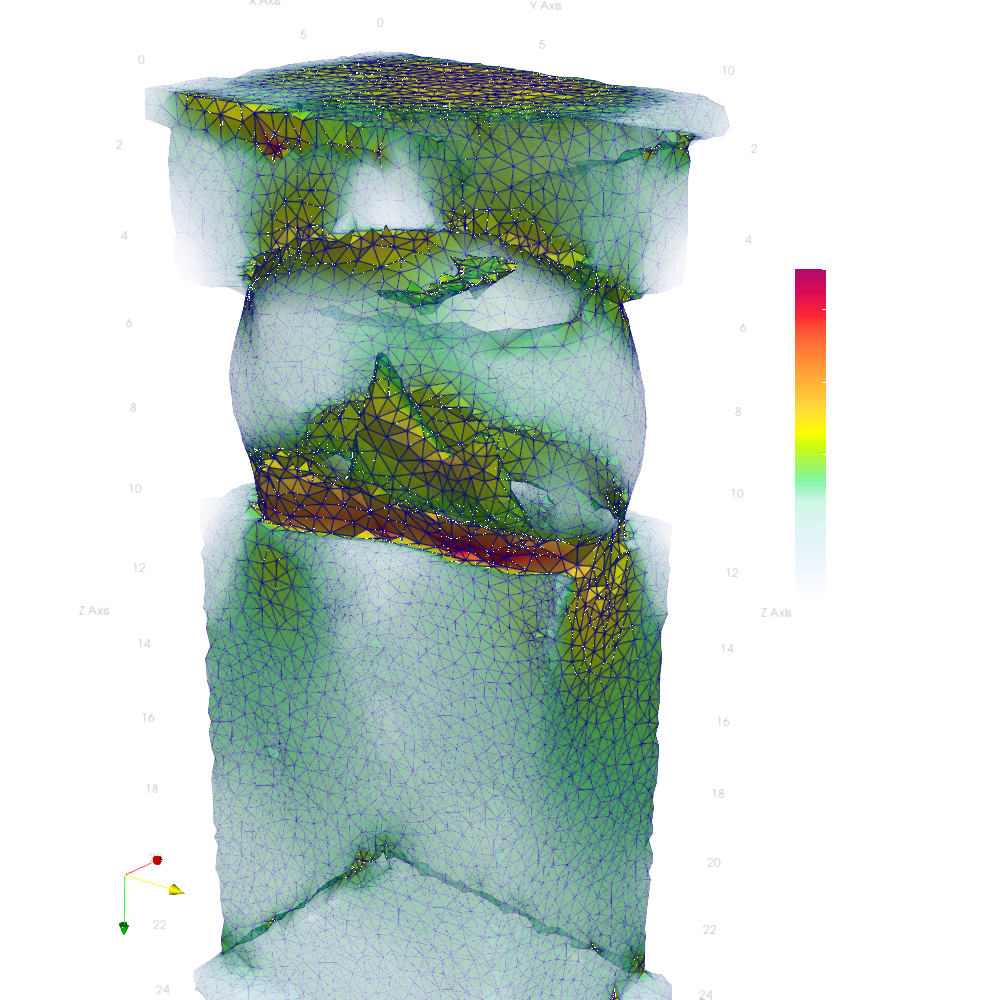
- Plot midplanes of the Von Mises stress
S_Mises
ciclope.utils.postprocess.paraview_plot("test_data/tooth/results/Tooth_3_scaled_2.vtk", slicenormal="xyz",
RepresentationType="Surface", Crinkle=False, ColorBy="S_Mises", Roll=90,
ImageResolution=[1024, 1024])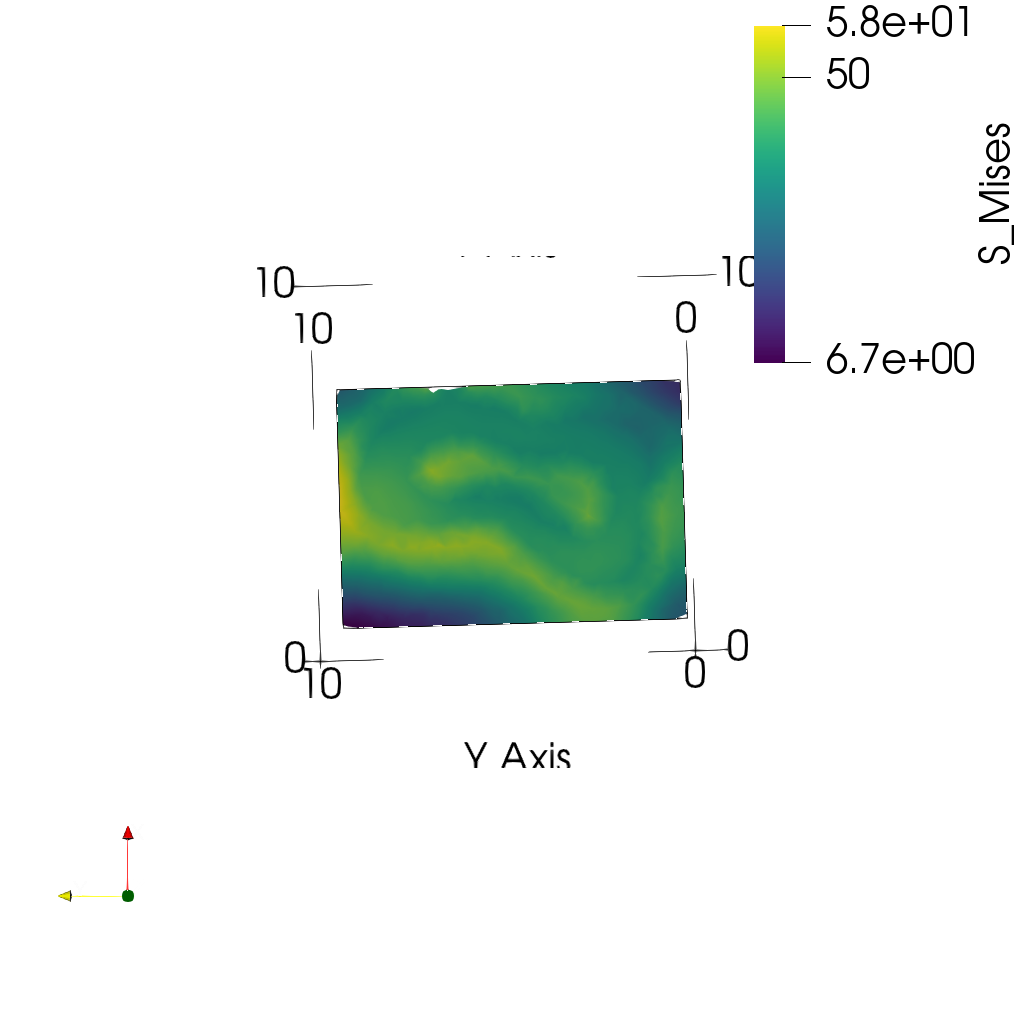 |
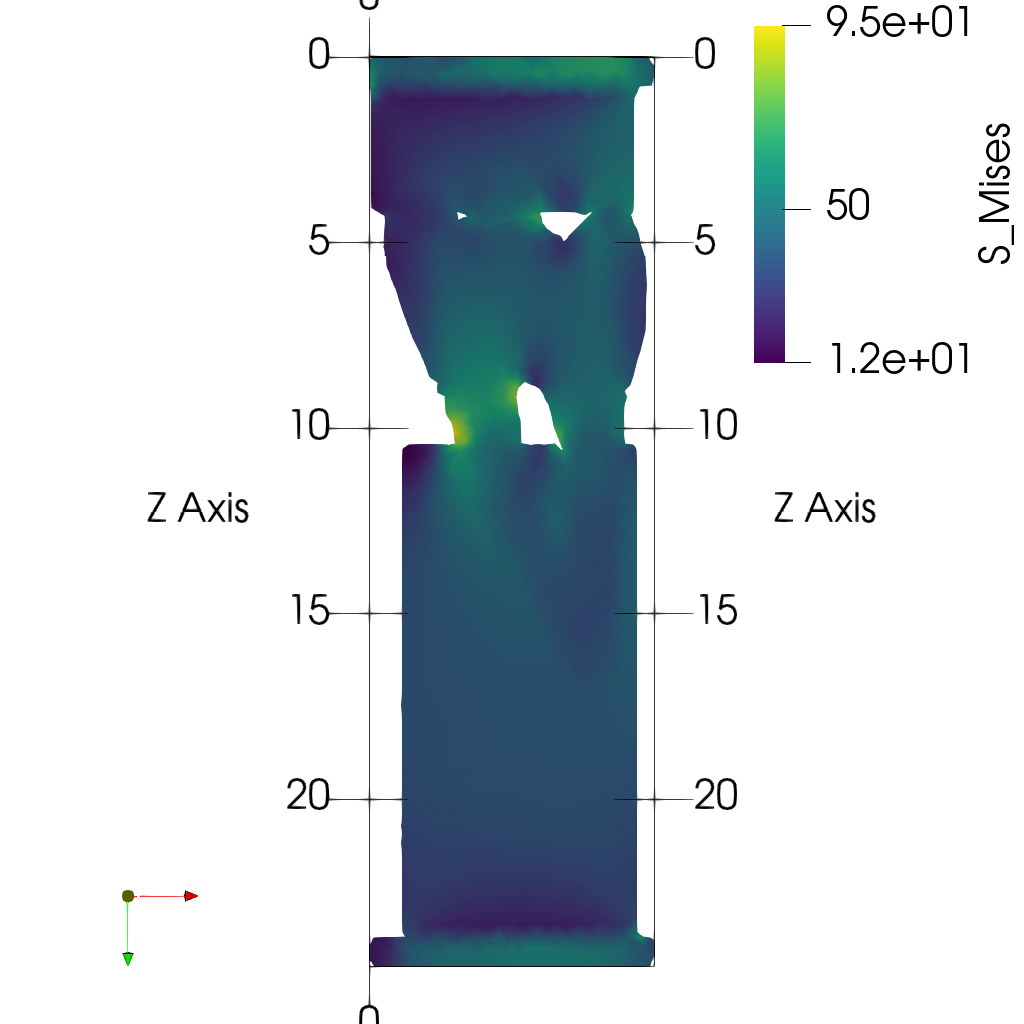 |
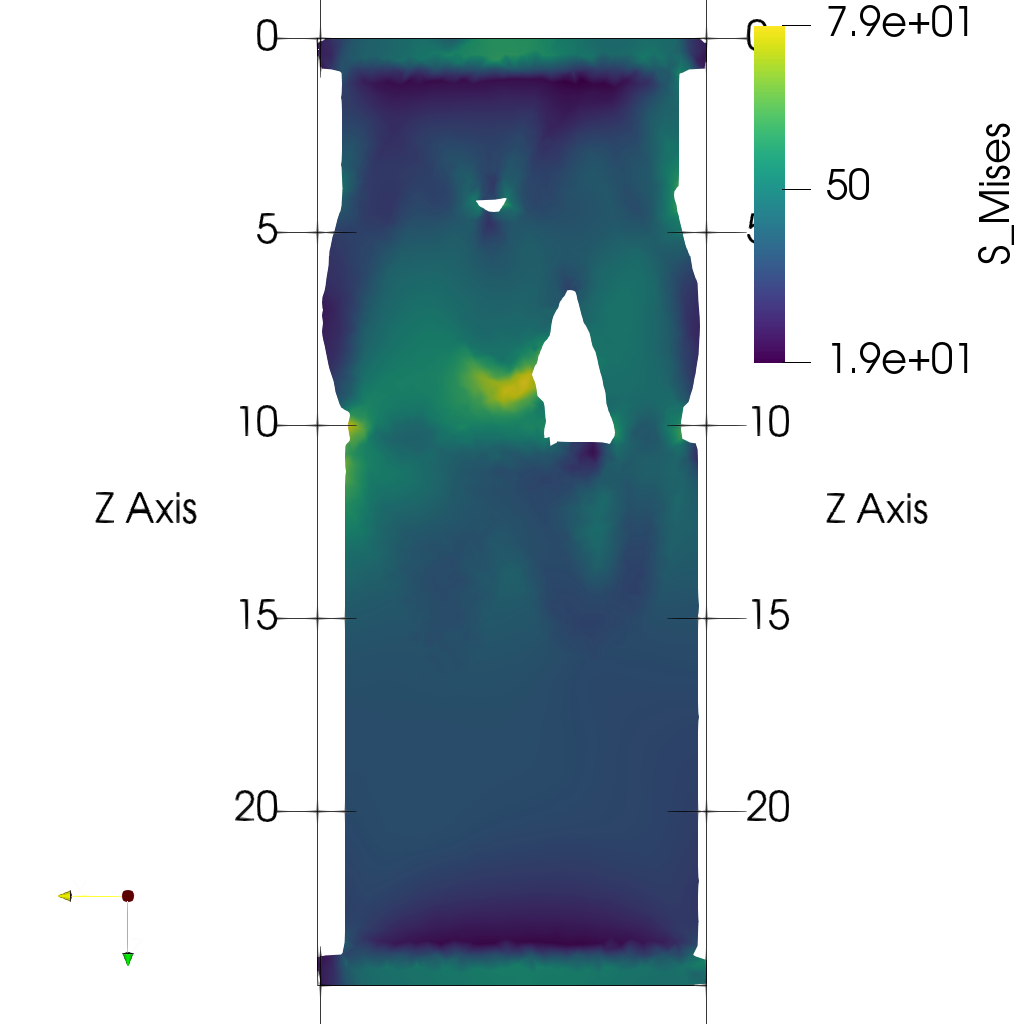 |
See the Jupyter Notebooks in the examples section for more examples of 3D data and results visualization.
The following table shows a general pipeline for FE model generation from CT data that can be executed from the command line with the ciclope script:
| # | Step | Description | command line script flag |
|---|---|---|---|
| 1. | Load CT data | ||
| 2. | Pre-processing | Gaussian smooth | --smooth |
| Resize image | -r |
||
| Add embedding | (not implemented yet) | ||
| Add caps | --caps |
||
| 3. | Segmentation | Uses Otsu method if left empty | -t |
| Remove unconnected voxels | |||
| 4. | Meshing | Outer shell mesh of triangles | --shell_mesh |
| Volume mesh of tetrahedra | --vol_mesh |
||
| 5. | FE model generation | Apply Boundary Conditions | |
| Material mapping | -m, --mapping |
||
| Voxel FE | --voxelfe |
||
| Tetrahedra FE | --tetrafe |
- Tetrahedra meshes are generated with pygalmesh (a Python frontend to CGAL)
- High-resolution surface meshes for visualization are generated with the PyMCubes module.
- All mesh exports are performed with the meshio module.
- ciclope handles the definition of material properties and FE analysis parameters (e.g. boundary conditions, simulation steps..) through separate template files. The folders material_properties and input_templates contain a library of template files that can be used to generate FE simulations.
The pipeline can be executed from the command line with:
ciclope test_data/LHDL/3155_D_4_bc/cropped/3155_D_4_bc_0000.tif test_data/LHDL/3155_D_4_bc/results/3155_D_4_bc_voxelFE.inp -vs 0.0195 0.0195 0.0195 -r 2 -t 63 --smooth 1 --voxelfe --template input_templates/tmp_example01_comp_static_bone.inp --verbose
The example shows how to:
- Load and inspect microCT volume data
- Apply Gaussian smooth
- Resample the dataset
- Segment the bone tissue
- Remove unconnected clusters of voxels
- Convert the 3D binary to a voxel-FE model for simulation in CalculX or Abaqus
- Linear, static analysis; displacement-driven
- Local material mapping (dataset Grey Values to bone Tissue Elastic Modulus)
- Launch simulation in Calculix
- Convert Calculix output to .VTK for visualization in Paraview
- Visualize simulation results in Paraview
The pipeline can be executed from the command line with:
ciclope test_data/LHDL/3155_D_4_bc/cropped/3155_D_4_bc_0000.tif test_data/LHDL/3155_D_4_bc/results/3155_D_4_bc.inp -vs 0.0195 0.0195 0.0195 -r 2 -t 63 --smooth 1 --tetrafe --max_facet_distance 0.025 --max_cell_circumradius 0.05 --vol_mesh --template input_templates/tmp_example01_comp_static_bone.inp
The pipeline can be executed from the command line with:
ciclope input.tif output.inp -vs 0.0065 0.0065 0.0065 --smooth -r 1.2 -t 90 --vol_mesh --tetrafe --template ./../input_templates/tmp_example02_tens_Nlgeom_steel.inp -v
The example shows how to:
- Load and inspect synchrotron microCT volume data
- Apply Gaussian smooth
- Resample the dataset
- Segment the steel
- Remove unconnected clusters of voxels
- Generate volume mesh of tetrahedra
- Generate high-resolution mesh of triangles of the model outer shell (for visualization)
- Convert the 3D binary to a tetrahedra-FE model for simulation in CalculX or Abaqus
- Non-linear, quasi-static analysis definition: tensile test with material plasticity. For more info visit: github.com/mkraska/CalculiX-Examples
- Local material mapping
- Launch simulation in Calculix
- Convert Calculix output to .VTK for visualization in Paraview
- Visualize simulation results in Paraview
This project was partially developed during the Jupyter Community Workshop “Building the Jupyter Community in Musculoskeletal Imaging Research” sponsored by NUMFocus.