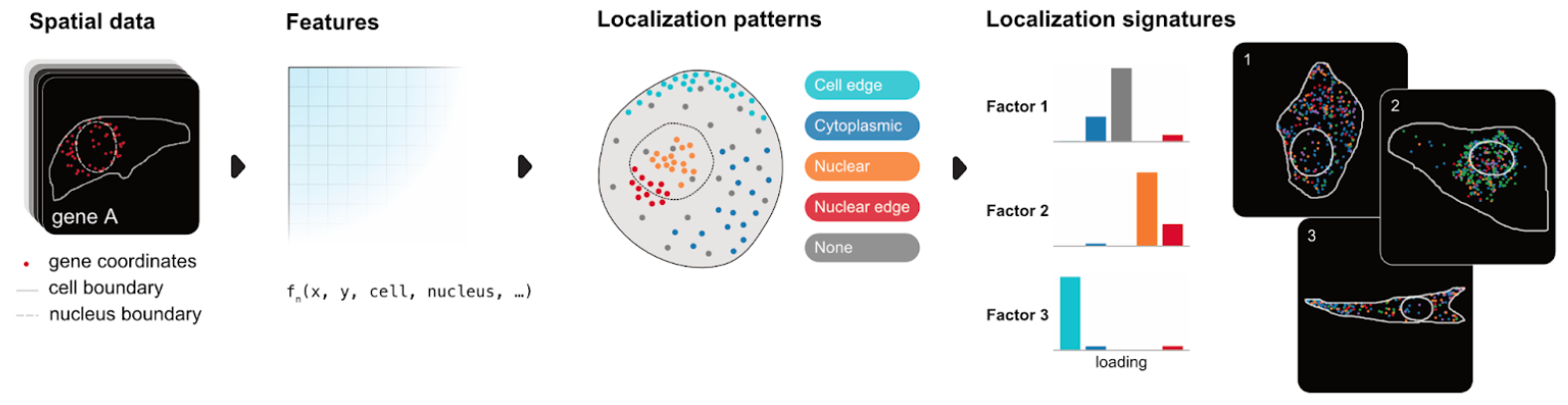
Bento is a Python toolkit for performing subcellular analysis of spatial transcriptomics data.
Install with Python 3.8 or 3.9:
pip install bento-toolsCheck out the documentation for the installation guide, tutorials, API and more! Read and cite our preprint ____ if you use Bento in your work.
- Store molecular coordinates and segmentation masks
- Visualize spatial transcriptomics data at subcellular resolution
- Compute subcellular spatial features
- Predict localization patterns and signatures
- Factor decomposition for high-dimensional spatial feature sets