This is the repo containing code for a section of the final chapter of my thesis which uses a simple model of cell-cell communication, known as Delta-Notch signalling to demonstrate concretely how local interactions between cells can improve their positional inference (or, in other words, how it can improve the coordination between the internal state of a cell and its position).
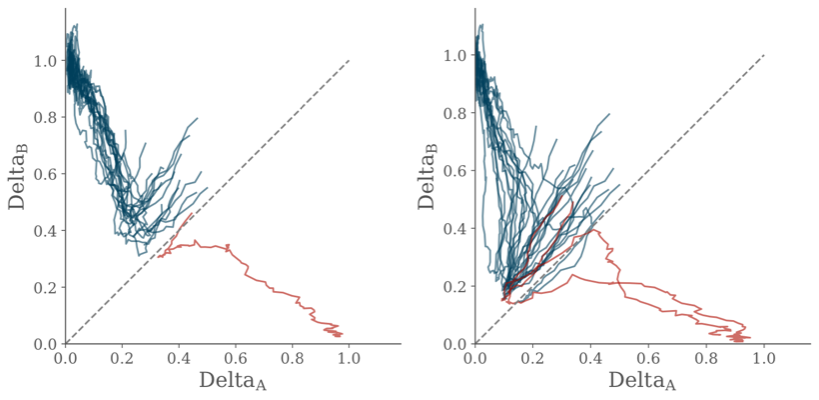
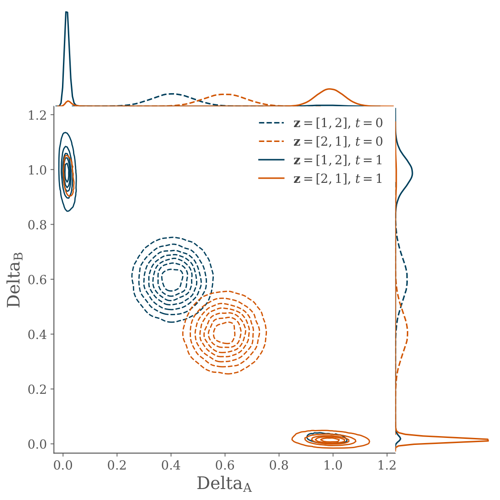
A simple two-cell model is used to illustrate the link between maximum-likelihood inference of position and the dynamical features of a gene-regulatory network.
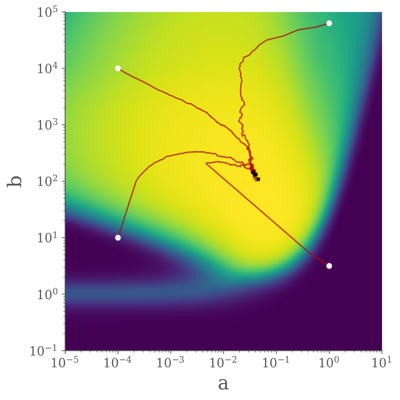
The parameters of the SDE are then optimised with respect to the system's performance on the patterning problem. This uses the autodiff functionality of Jax so take gradients through the (somewhat complex) objective function and subsequently the SDE solver.
The Delta-Notch system is modelled using the equations of Collier et al. (1996) which form the basis of the stochastic differential equations.
The code uses the jax-based SDE solvers from xwinxu/bayeSDE.
This repo has become a bit messy during the somewhat hectic final stages of writing my thesis. It will be repaired shortly.
While sde solvers in the bayeSDE repo have worked well, ideally this code would now be converted to a more fully fledged Jax-based differential equation library such as patrick-kidger/diffrax.