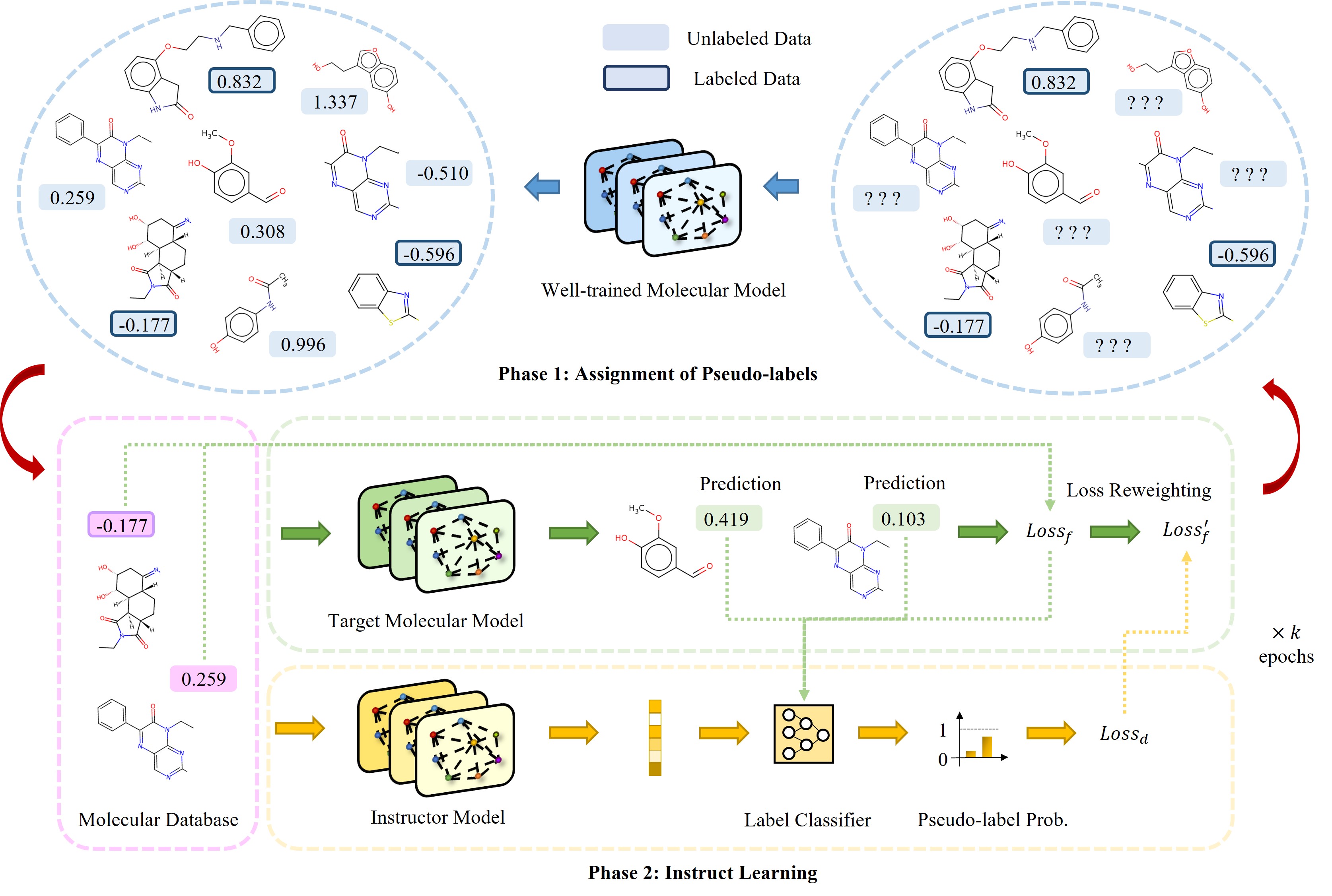
This is the official repository for the paper InstructBio: A Large-scale Semi-supervised Learning Paradigm for Biochemical Problems
[arXiv].
We propose a novel and efficient semi-supervised learning framework to overcome the limitation of labeled data scarcity and the difficulty of
annotating molecular data.
We conduct extensive experiments in MoleculeNet [1] and MoleculeACE [2] to verify the efficacy of our InstructBio and further results show that
InstructBio can be collaborated with pretrained molecular models such as GEM [3] to achieve better performance. Besides, we validate that
we can adopt molecular models trained by InstructBio to build large-scale and task-specific pseudo-labeled molecular datasets, which can
be reused to train other kinds of molecular models and obtain a more powerful representation capability.
Table of Contents
InstructBio currently supports Python 3.7. We rely on PyTorch (1.10.0) and
PyTorch Geometric (2.2.0) to execute the code.
Notably, please do not install MoleculeACE by pip install MoleculeACE, because we make necessary modifications on its original implementation.
Please follow the instruction below to install the necessary environment.
pip install torch # we use torch 1.10.0 & cu113
pip install -U scikit-learn
pip install torch-cluster -f https://data.pyg.org/whl/torch-1.10.0+cu113.html
pip install torch-scatter -f https://data.pyg.org/whl/torch-1.10.0+cu113.html
pip install torch-sparse -f https://data.pyg.org/whl/torch-1.10.0+cu113.html
pip install torch-geometricBefore implementation of InstructBio, we need to acquire both the labeled and unlabeled molecular data. In the main text, we select two categories of problems: molecular property prediction and the activity cliff estimation.
Unlabeled data are collected from the common database ZINC15, a free database of commercially-available compounds for virtual screening. ZINC15 contains over 230 million purchasable compounds in ready-to-dock, 3D formats. You can use DeepChem to attain the unlabeled SMILES of molecules via this following script:
https://github.com/deepchem/deepchem/blob/master/deepchem/molnet/load_function/zinc15_datasets.pyThere are four different sizes of ZINC15: 250K, 1M, 10M, and 270M. We only utilize the 250K and 1M versions.
We adopt the MoleculeNet [1] as the benchmark to evaluate our model for molecular property prediction, where 3 regression tasks and 6 classification tasks are selected. The data can be obtained from either the official website of MoleculeNet at this link or from GEM [3] by running the following line:
wget https://baidu-nlp.bj.bcebos.com/PaddleHelix/datasets/compound_datasets/chemrl_downstream_datasets.tgz
Notably, before using them, you need to rename the file names rather than putting them into the folder MoleculeACE/benchmark/Data/benchmark_data.
The name dictionary is listed as follows: ['freesolv', 'esol', 'lipo', 'bbbp', 'bace', 'clintox', 'tox21', 'toxcast', 'sider'].
For convenience, we have provided preprocessed MoleculeNet datasets in this repository, and you can directly run relevant tasks.
We employ a scaffold splitting to split the dataset into training/validation/test sets. The splitter can be found at MoleculeACE/benchmark/utils.py.
We use the MoleculeACE [2] benchmark to validate the capability of our InstructBio in addressing the activity cliff estimation. MoleculeACE contains
30 datasets, each corresponding to a target macromolecule. Its data is available at its official GitHub at this
URL, where data splits are already provided.
Then you need to put the data (in .csv) under the folder MoleculeACE/benchmark/Data/benchmark_data and
we use a stratified scaffold splitting to evaluate the performance of different GNNs.
In order to realize the semi-supervised learning of our InstrutBio, please run the following line of code:
python main.py --model=GIN --data=xxxxxx --critic_epochs=20 --update_label_freq=5 where you can change the model type from ['GCN', 'GIN', 'GAT']. You can also choose the dataset by altering --data arguement from
['CHEMBL4203_Ki', 'CHEMBL2034_Ki', 'CHEMBL233_Ki', 'CHEMBL4616_EC50', 'CHEMBL287_Ki', 'CHEMBL218_EC50', 'CHEMBL264_Ki', 'CHEMBL219_Ki', 'CHEMBL2835_Ki', 'CHEMBL2147_Ki', 'CHEMBL231_Ki', 'CHEMBL3979_EC50', 'CHEMBL237_EC50', 'CHEMBL244_Ki', 'CHEMBL4792_Ki', 'CHEMBL1871_Ki', 'CHEMBL237_Ki', 'CHEMBL262_Ki', 'CHEMBL2047_EC50', 'CHEMBL239_EC50', 'CHEMBL2971_Ki', 'CHEMBL204_Ki', 'CHEMBL214_Ki', 'CHEMBL1862_Ki', 'CHEMBL234_Ki', 'CHEMBL238_Ki', 'CHEMBL235_EC50', 'CHEMBL4005_Ki', 'CHEMBL236_Ki', 'CHEMBL228_Ki']. You can control the number of
unlabeled data size via --unlabeled_size to use only a portion of all available unlabeled data.
You can also add an extra commend --debug to only run one fold for debugging. Many other kinds of arguments are supported as well and please
check the code for details. Notably, in order to fast up the training speed for different tasks with the same unlabeled database, we save the processed unlabeled data file the
first time reading these unlabeled data points.
We can also use a molecular model well-trained by InstructBio to produce a hybrid dataset and use that hybrid dataset to train other deep learning architectures. This can be done by running the following code:
python teacher_main.py --model=GIN --teacher_model=GIN --data=xxxxxx --normalize=1 where you need to specify the type of the student model and the teacher model (e.g., the one to annotate the unlabeled corpus).
If you find our helpful and interesting, please consider citing our paper. Thank you! 😜 Any kind of question is welcome, and you can directly pull an issue or send emails to Fang WU. We are pleased to see future works based on our InstructBio to push the frontier of molecular representation learning through semi-supervised learning.
@article{wu2023instructbio,
title={InstructBio: A Large-scale Semi-supervised Learning Paradigm for Biochemical Problems},
author={Wu, Fang and Qin, Huiling and Gao, Wenhao and Li, Siyuan and Coley, Connor W and Li, Stan Z and Zhan, Xianyuan and Xu, Jinbo},
journal={arXiv preprint arXiv:2304.03906},
year={2023}
}InstructBio is under MIT license. For use of specific models, please refer to the model licenses found in the original packages.
[1]
Wu, Zhenqin, et al. "MoleculeNet: a benchmark for molecular machine learning." Chemical Science 9.2 (2018): 513-530.
[2]
van Tilborg, Derek, Alisa Alenicheva, and Francesca Grisoni. "Exposing the limitations of molecular machine learning with activity cliffs."
Journal of Chemical Information and Modeling 62.23 (2022): 5938-5951.
[3]
Fang, Xiaomin, et al. "Geometry-enhanced molecular representation learning for property prediction." Nature Machine Intelligence 4.2 (2022): 127-134.