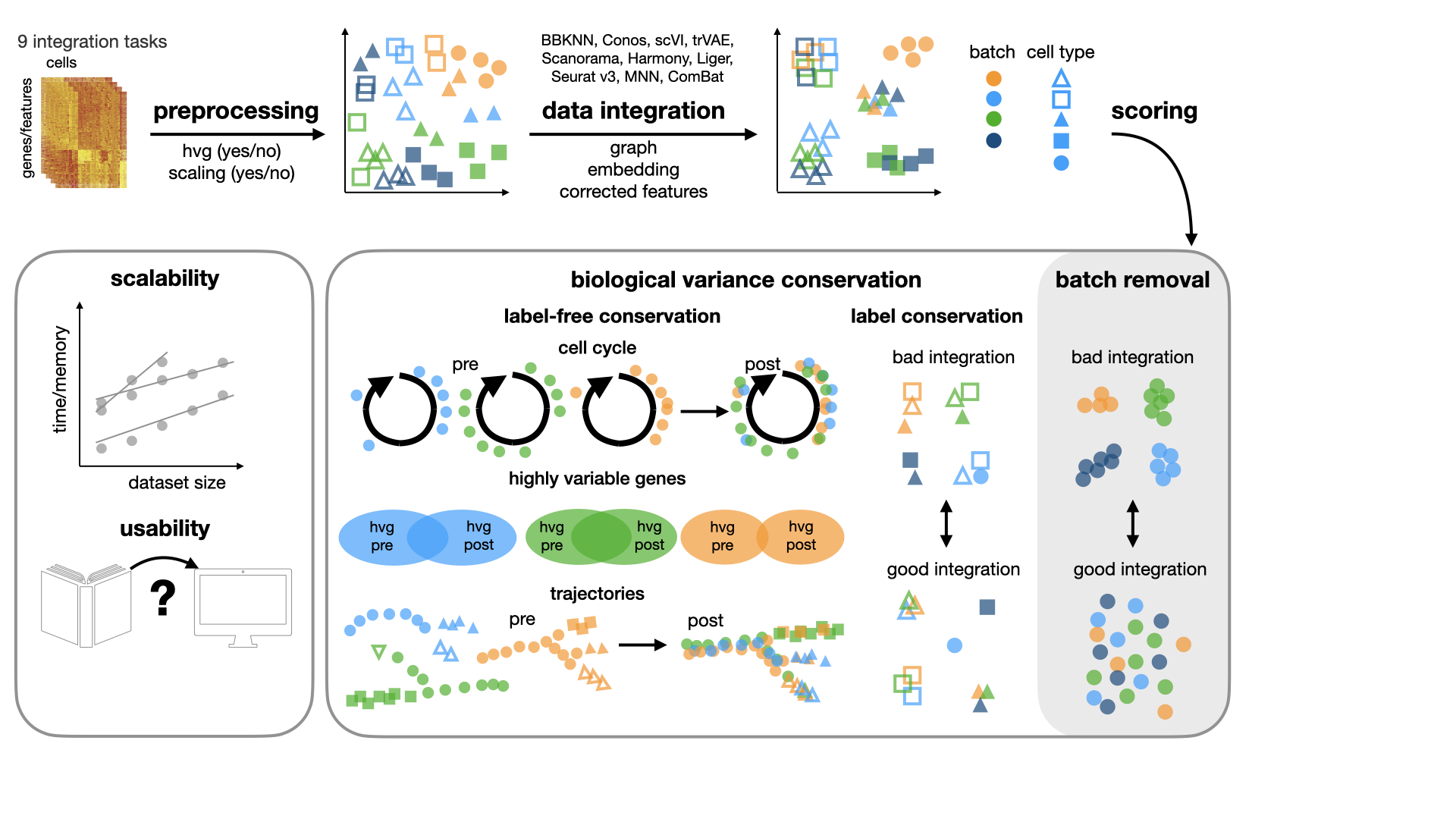
This repository contains the code for our benchmarking study for data integration tools. In our study, we benchmark 16 methods (see here) with 4 combinations of preprocessing steps leading to 68 methods combinations on 85 batches of gene expression and chromatin accessibility data.
-
On our website we visualise the results of the study.
-
The reusable pipeline we used in the study can be found in the separate scIB pipeline repository. It is reproducible and automates the computation of preprocesssing combinations, integration methods and benchmarking metrics.
-
For reproducibility and visualisation we have a dedicated repository: scib-reproducibility.
Benchmarking atlas-level data integration in single-cell genomics. MD Luecken, M Büttner, K Chaichoompu, A Danese, M Interlandi, MF Mueller, DC Strobl, L Zappia, M Dugas, M Colomé-Tatché, FJ Theis bioRxiv 2020.05.22.111161; doi: https://doi.org/10.1101/2020.05.22.111161_
We created the python package called scIB that uses scanpy to streamline the integration of single-cell datasets and
evaluate the results. The evaluation of integration quality is based on a number of metrics.
The scIB python package is in the folder scIB. You can install it from the root of this repository using
pip install .
Alternatively, you can also install the package directly from GitHub via
pip install git+https://github.com/theislab/scib.git
Additionally, in order to run the R package kBET, you need to install it through R.
devtools::install_github('theislab/kBET')We recommend to use a conda environment or something similar, so that python and R dependencies are in one place. Please also check out scIB pipeline for ready-to-use environments.
This package contains code for running integration methods as well as for evaluating their output. However, due to
dependency clashes, scIB is only installed with the packages needed for the metrics. In order to use the integration
wrapper functions, we recommend to work with different environments for different methods, each with their own
installation of scIB. Check out the Tools section for a list of supported integration methods.
The package contains several modules for the different steps of the integration and benchmarking pipeline. Functions for
the integration methods are in scIB.integration. The methods can be called using
scIB.integration.run<method>(adata, batch=<batch>)
where <method> is the name of the integration method and <batch> is the name of the batch column in adata.obs.
Some integration methods (scGEN, SCANVI) also use cell type labels as input. For these, you need to additionally provide the corresponding label column.
runScGen(adata, batch=<batch>, cell_type=<cell_type>)
runScanvi(adata, batch=<batch>, labels=<cell_type>)
scIB.preprocessing contains methods for preprocessing of the data such as normalisation, scaling or highly variable
gene selection per batch. The metrics are located at scIB.metrics. To run multiple metrics in one run, use
the scIB.metrics.metrics() function.
For a detailed description of the metrics implemented in this package, please see the manuscript.
- Principal component regression
pcr_comparison() - Batch ASW
silhouette() - K-nearest neighbour batch effect
kBET() - Graph connectivity
graph_connectivity() - Graph iLISI
lisi_graph()
- Normalised mutual information
nmi() - Adjusted Rand Index
ari() - Cell type ASW
silhouette_batch() - Isolated label score F1
isolated_labels() - Isolated label score ASW
isolated_labels() - Cell cycle conservation
cell_cycle() - Highly variable gene conservation
hvg_overlap() - Trajectory conservation
trajectory_conservation() - Graph cLISI
lisi_graph()
Tools that are compared include: