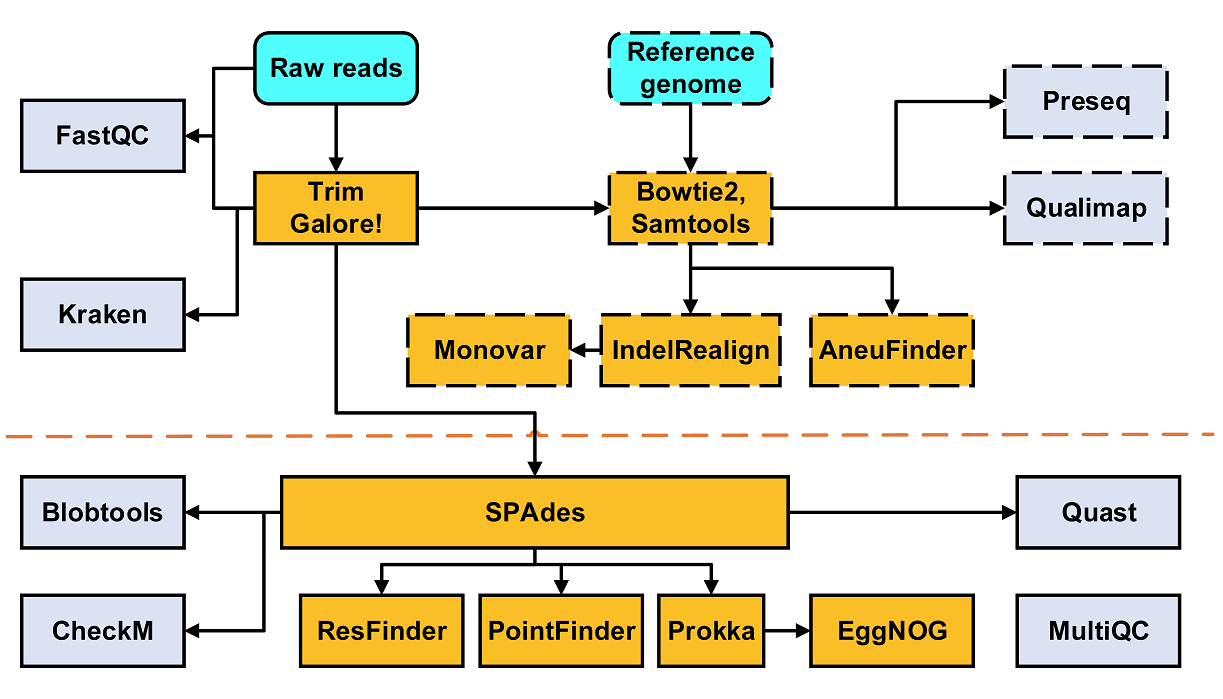
Single Cell Genome Sequencing data analysis pipeline.
The pipeline is used for single cell genome sequencing data analysis and built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker / singularity containers making installation trivial and results highly reproducible.
Prerequisites: Git, Java 11 or later, Docker
## Install Nextflow
$ curl -s https://get.nextflow.io | bash
## Get the pipeline
$ git clone -b v2.0.1 https://github.com/gongyh/nf-core-scgs.git
## Test
$ ./nextflow run nf-core-scgs -profile test_local,docker
or $ ./nextflow run nf-core-scgs -profile test_local,podman
or $ APPTAINER_DISABLE_CACHE=true ./nextflow run nf-core-scgs -profile test_local,apptainer
or $ SINGULARITY_DISABLE_CACHE=true ./nextflow run nf-core-scgs -profile test_local,singularity
or $ ./nextflow run nf-core-scgs -profile test_local,conda
# for conda, add `disable_lockfile: true` to ~/.condarc or ~/.mambarcThe gongyh/nf-core-scgs pipeline comes with documentation about the pipeline, found in the docs/ directory:
- Installation
- Pipeline configuration
- Running the pipeline
- Output and how to interpret the results
- Troubleshooting
[1] Jing, X., Gong, Y., Pan, H. et al. Single-cell Raman-activated sorting and cultivation (scRACS-Culture) for assessing and mining in situ phosphate-solubilizing microbes from nature. ISME COMMUN. 2, 106 (2022). https://doi.org/10.1038/s43705-022-00188-3
[2] Jing X, Gong Y, Xu T, Meng Y, Han X, Su X, Wang J, Ji Y, Li Y, Jia Z, Ma B, Xu J. One-Cell Metabolic Phenotyping and Sequencing of Soil Microbiome by Raman-Activated Gravity-Driven Encapsulation (RAGE). mSystems. 2021 Jun 29;6(3):e0018121. doi: 10.1128/mSystems.00181-21. Epub 2021 May 27. PMID: 34042466; PMCID: PMC8269212
[3] Xu, T., Gong, Y., Su, X., Zhu, P., Dai, J., Xu, J., Ma, B., Phenome-Genome Profiling of Single Bacterial Cell by Raman-Activated Gravity-Driven Encapsulation and Sequencing. Small 2020, 2001172. https://doi.org/10.1002/smll.202001172
[4] Su, X., Gong, Y., Gou, H., Jing, X., Xu, T., Zheng, X., Chen, R., Li, Y., Ji, Y., Ma, B., Xu, J., Rational Optimization of Raman-Activated Cell Ejection and Sequencing for Bacteria. Analytical Chemistry, 2020. https://doi.org/10.1021/acs.analchem.9b05345
gongyh/nf-core-scgs is developed by Yanhai Gong, maintained by Yanhai Gong and Shiqi Zhou. We look forward to receive your feedback, bug reports, or suggestions for the further development of this pipeline.
This pipeline is open source under the MIT license, and integrates wonderful third-party softwares, which remain owned and copyrighted by their respective developers. Authors cannot be held legally or morally responsible for any consequences that may arise from using or misusing it.