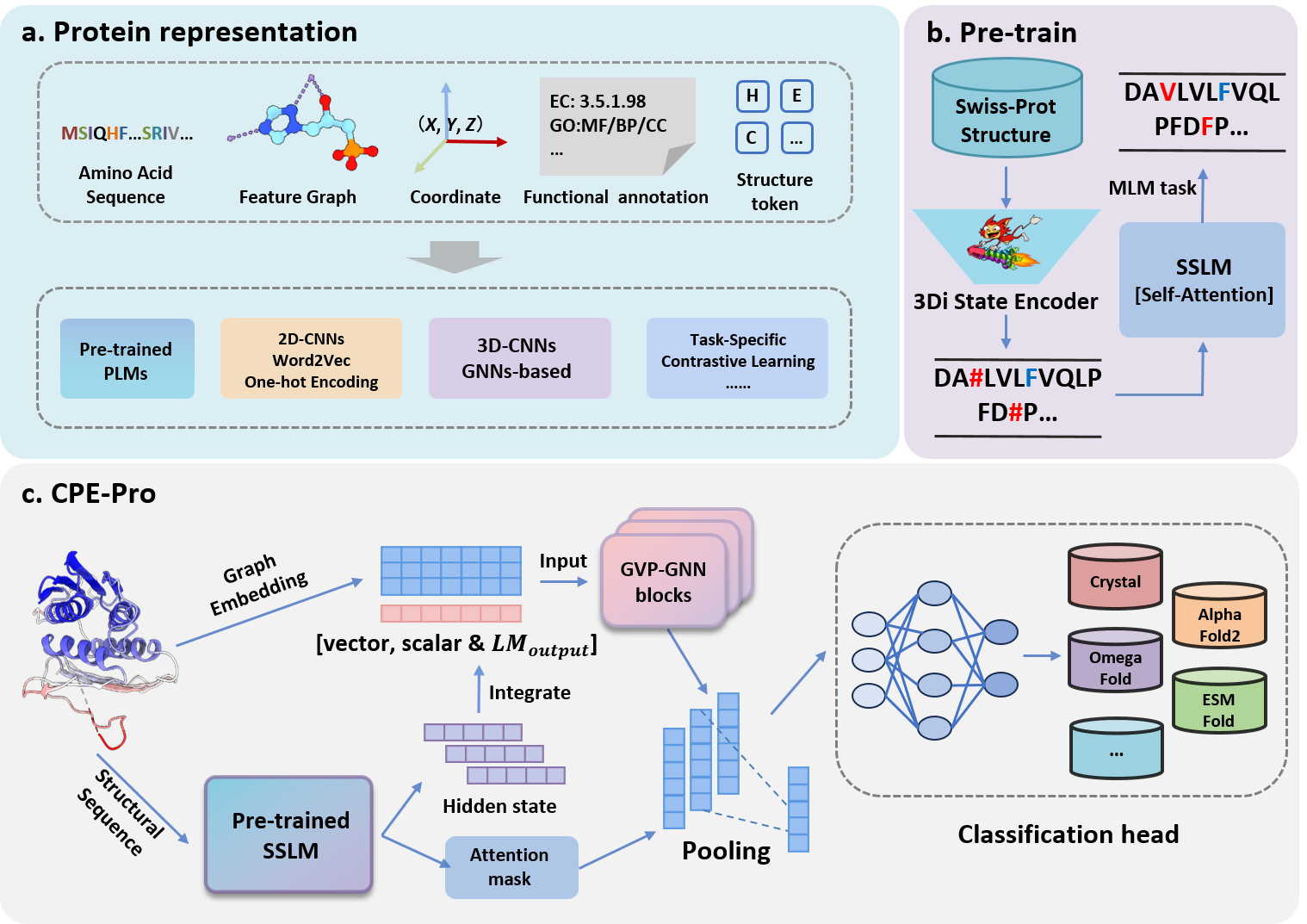
CPE-Pro: A structure-sensitive supervised deep learning model, Crystal vs Predicted Evaluator for Protein Structure, to represent and discriminate the origin of protein structures. CPE-Pro integrates two distinct structure encoders corresponding to graphical and sequential representations of the structures.
The sequential encoder of CPE-Pro is the Structural Sequence Language Model (SSLM). First, the protein structure data search tool Foldseek is used to convert protein structures into "structure-sequences". Next, using the 3Di alphabet as the vocabulary for structural elements and based on the Transformer architecture, we pre-train a protein structural language model, SSLM, from scratch. This aims to effectively model the "structure-sequences" of proteins. The pre-training process employs the classic masked language modeling (MLM) objective, predicting masked elements based on the context of the "structure-sequences".
We compared CPE-Pro to various embedded-based deep learning methods on the dataset CATH-PFD. Our analysis includes pre-trained PLMs(ESM1b, ESM1v, ESM2, ProtBert) combined with GVP-GNN as a model with amino acid sequence and structure input, and the structure-aware PLM SaProt.
(1) Results show CPE-Pro demonstrates exceptionally high accuracy performance in two structure discrimination tasks (C-A: Crystal - AlphaFold2, C-M: Crystal - Multiple prediction models).
(2) Feature Visualization Method t-SNE Powerfully Demonstrates pretrained SSLM’s Excellence in Capturing Structural Differences.
(3) Preliminary experiments indicate that, compared to amino acid sequences, "structure-sequences" enable language models to learn more effective protein feature information, enriching and optimizing structural representations when integrated with graph embeddings.
Please make sure you have installed Anaconda3 or Miniconda3.
Then
cd CPE-Pro-main
You can create the required environment using the following two methods.
conda env create -f environment.yaml
conda activate cpe-pro
or
conda create -n cpe-pro python=3.8.18
conda activate cpe-pro
pip install -r requirements.txt
All protein folding, pre-training, and experiments were conducted on 8 NVIDIA RTX 3090 GPUs. If you intend to utilize a larger pre-trained SSLM or a deeper GVP-GNN within CPE-Pro, additional hardware resources may be necessary.
The link to the dataset can be found in the source folder.
(1) Swiss-Prot: The pre-training of the protein structural sequence language model, SSLM utilized 109,334 high pLDDT score (>0.955) protein structures from the Swiss-Prot database. We organized the protein "structure-sequences" used in the pre-training process into a FASTA file.
(2) CATH: Dataset cath-dataset-nonredundant-S40-v4_3_0.pdb as our benchmark, then we extracted the AA sequences of proteins from the benchmark dataset. Using multiple state-of-the-art protein structure prediction models, we predicted the structures corresponding to these AA sequences. These structures were organized and categorized based on individual proteins and prediction models to construct a Protein Folding Dataset, CATH-PFD, which will be used for training and validating CPE-Pro.
(3) SCOPe: We selected a subset of gene domain sequences from the non-redundant Astral SCOPe 2.08 database in SCOPe, where the identity between sequences is less than 40%. From this subset, we focused on all-α helical proteins (2,644) and all-β sheet proteins (3,059) and filtered the corresponding structural sets in the database.
(4) Case Study(BLAT ECOLX and CP2C9 HUMAN): In three structural prediction models, both proteins achieved pLDDT scores above 0.9, indicating high accuracy in structure prediction with minimal deviation from the crystal structure.
The information about the model's weight files can be found in the source folder.
(1) See the pretrain.py script for pretraining details. Examples can be found in pretrain folder.
(2) See the train.py script for training details. Examples can be found in scripts folder.
Please cite our work if you have used our code or data.
xxxx