Foldseek enables fast and sensitive comparisons of large protein structure sets.
- Foldseek
- Table of Contents
Search your protein structures against the AlphaFoldDB and PDB in seconds using the Foldseek webserver (code): search.foldseek.com 🚀
# Linux AVX2 build (check using: cat /proc/cpuinfo | grep avx2)
wget https://mmseqs.com/foldseek/foldseek-linux-avx2.tar.gz; tar xvzf foldseek-linux-avx2.tar.gz; export PATH=$(pwd)/foldseek/bin/:$PATH
# Linux SSE2 build (check using: cat /proc/cpuinfo | grep sse2)
wget https://mmseqs.com/foldseek/foldseek-linux-sse2.tar.gz; tar xvzf foldseek-linux-sse2.tar.gz; export PATH=$(pwd)/foldseek/bin/:$PATH
# Linux ARM64 build
wget https://mmseqs.com/foldseek/foldseek-linux-arm64.tar.gz; tar xvzf foldseek-linux-arm64.tar.gz; export PATH=$(pwd)/foldseek/bin/:$PATH
# MacOS
wget https://mmseqs.com/foldseek/foldseek-osx-universal.tar.gz; tar xvzf foldseek-osx-universal.tar.gz; export PATH=$(pwd)/foldseek/bin/:$PATH
# Conda installer (Linux and macOS)
conda install -c conda-forge -c bioconda foldseek
Other precompiled binaries for ARM64 amd SSE2 are available at https://mmseqs.com/foldseek.
For optimal software performance, consider three options based on your RAM and search requirements:
-
With Cα info (default). Use this formula to calculate RAM -
(6 bytes Cα + 1 3Di byte + 1 AA byte) * (database residues). The 54M AFDB50 entries require 151GB. -
Without Cα info. By disabling
--sort-by-structure-bits 0, RAM requirement reduces to 35GB. However, this alters hit rankings and final scores but not E-values. Structure bits are mostly relevant for hit ranking for E-value > 10^-1. -
Single query searches. Use the
--prefilter-mode 1, which isn't memory-limited and computes all ungapped alignments. This option optimally utilizes foldseek's multithreading capabilities for single queries.
We presented a Foldseek tutorial at the SBGrid where we demonstrated Foldseek's webserver and command line interface. Check it out here.
Many of Foldseek's modules (subprograms) rely on MMseqs2. For more information about these modules, refer to the MMseqs2 wiki. For documentation specific to Foldseek, checkout the Foldseek wiki here.
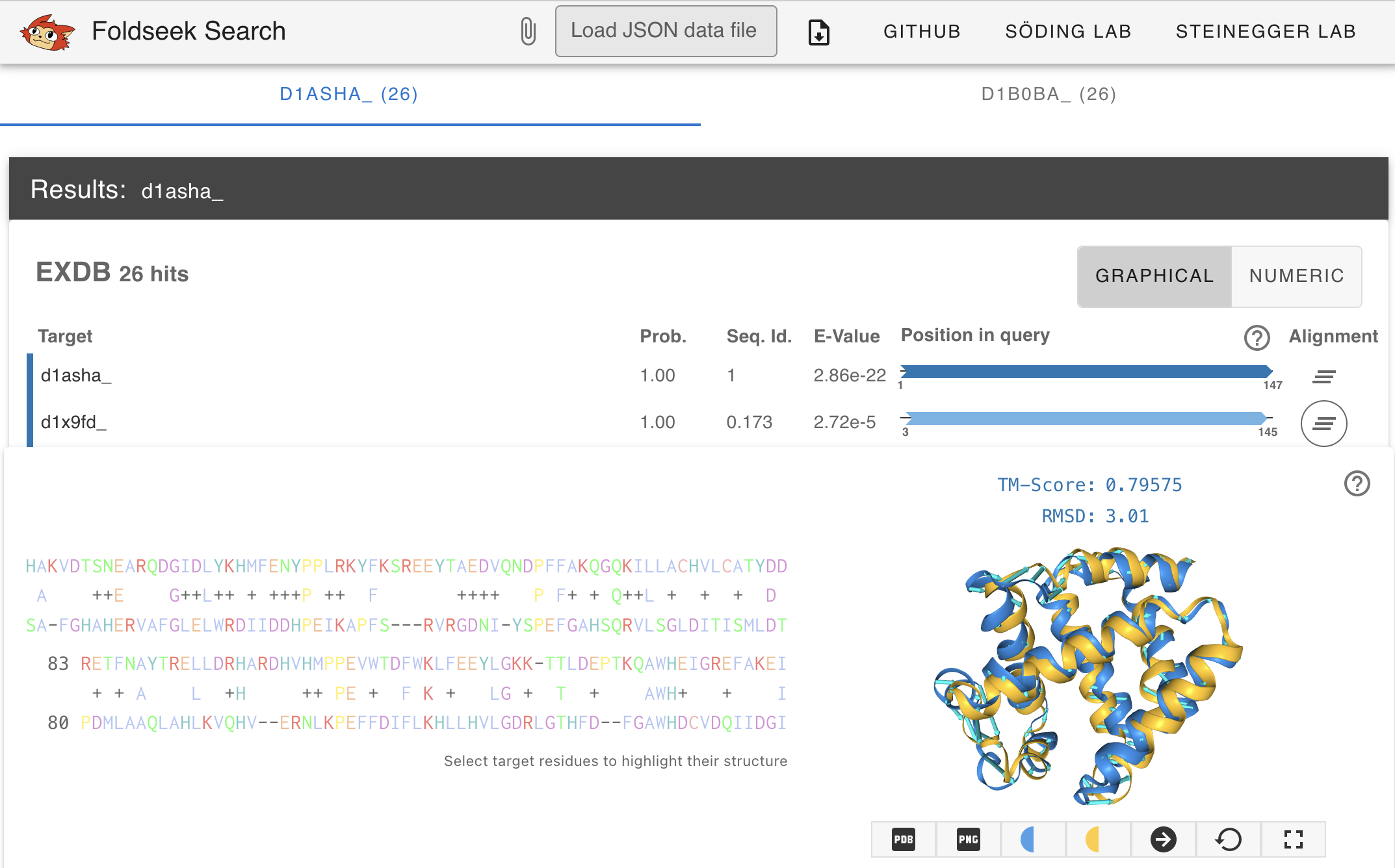
The easy-search module allows to query one or more single-chain protein structures, formatted in PDB/mmCIF format (flat or gzipped), against a target database, folder or individual single-chain protein structures (for multi-chain proteins see complexsearch). The default alignment information output is a tab-separated file but Foldseek also supports Superposed Cα PDBs and HTML.
foldseek easy-search example/d1asha_ example/ aln tmpFolder
The default output fields are: query,target,fident,alnlen,mismatch,gapopen,qstart,qend,tstart,tend,evalue,bits but they can be customized with the --format-output option e.g., --format-output "query,target,qaln,taln" returns the query and target accessions and the pairwise alignments in tab-separated format. You can choose many different output columns.
| Code | Description |
|---|---|
| query | Query sequence identifier |
| target | Target sequence identifier |
| qca | Calpha coordinates of the query |
| tca | Calpha coordinates of the target |
| alntmscore | TM-score of the alignment |
| qtmscore | TM-score normalized by the query length |
| ttmscore | TM-score normalized by the target length |
| u | Rotation matrix (computed to by TM-score) |
| t | Translation vector (computed to by TM-score) |
| lddt | Average LDDT of the alignment |
| lddtfull | LDDT per aligned position |
| prob | Estimated probability for query and target to be homologous (e.g. being within the same SCOPe superfamily) |
Check out the MMseqs2 documentation for additional output format codes.
Foldseek's --format-mode 5 generates PDB files with all target Cα atoms superimposed onto the query structure based on the aligned coordinates.
For each pairwise alignment it will write its own PDB file, so be careful when using this options for large searches.
Locally run Foldseek can generate an HTML search result, similar to the one produced by the webserver by specifying --format-mode 3
foldseek easy-search example/d1asha_ example/ result.html tmp --format-mode 3
| Option | Category | Description |
|---|---|---|
| -s | Sensitivity | Adjust sensitivity to speed trade-off; lower is faster, higher more sensitive (fast: 7.5, default: 9.5) |
| --exhaustive-search | Sensitivity | Skips prefilter and performs an all-vs-all alignment (more sensitive but much slower) |
| --max-seqs | Sensitivity | Adjust the amount of prefilter handed to alignment; increasing it can lead to more hits (default: 1000) |
| -e | Sensitivity | List matches below this E-value (range 0.0-inf, default: 0.001); increasing it reports more distant structures |
| --alignment-type | Alignment | 0: 3Di Gotoh-Smith-Waterman (local, not recommended), 1: TMalign (global, slow), 2: 3Di+AA Gotoh-Smith-Waterman (local, default) |
| -c | Alignment | List matches above this fraction of aligned (covered) residues (see --cov-mode) (default: 0.0); higher coverage = more global alignment |
| --cov-mode | Alignment | 0: coverage of query and target, 1: coverage of target, 2: coverage of query |
By default, Foldseek uses its local 3Di+AA structural alignment but it also supports realigning hits using the global TMalign as well as rescoring alignments using TMscore.
foldseek easy-search example/d1asha_ example/ aln tmp --alignment-type 1
If alignment type is set to tmalign (--alignment-type 1), the results will be sorted by the TMscore normalized by query length. The TMscore is used for reporting two fields: the e-value=(qTMscore+tTMscore)/2 and the score=(qTMscore*100). All output fields (e.g., pident, fident, and alnlen) are calculated based on the TMalign alignment.
Search by predicting 3Di directly from amino acid sequences without the need for existing protein structures. This feature uses the ProstT5 protein language model and runs by default on CPU and is about 400-4000x compared to predicted structures by ColabFold.
foldseek databases ProstT5 weights tmp
foldseek databases PDB pdb tmp
foldseek easy-search QUERY.fasta pdb result.m8 tmp --prostt5-model weights
Or create your a structural database from a fasta files.
foldseek createdb db.fasta db --prostt5-model weights
Faster inference using GPU/CUDA is also supported. Compile from source with cmake -DCMAKE_BUILD_TYPE=Release -DENABLE_CUDA=1 -DCUDAToolkit_ROOT=Path-To-Cuda-Toolkit and call with createdb/easy-search --prostt5-model weights --gpu 1.
The databases command downloads pre-generated databases like PDB or AlphaFoldDB.
# pdb
foldseek databases PDB pdb tmp
# alphafold db
foldseek databases Alphafold/Proteome afdb tmp
We currently support the following databases:
Name Type Taxonomy Url
- Alphafold/UniProt Aminoacid yes https://alphafold.ebi.ac.uk/
- Alphafold/UniProt50 Aminoacid yes https://alphafold.ebi.ac.uk/
- Alphafold/Proteome Aminoacid yes https://alphafold.ebi.ac.uk/
- Alphafold/Swiss-Prot Aminoacid yes https://alphafold.ebi.ac.uk/
- ESMAtlas30 Aminoacid - https://esmatlas.com
- PDB Aminoacid yes https://www.rcsb.org
The target database can be pre-processed by createdb. This is useful when searching multiple times against the same set of target structures.
foldseek createdb example/ targetDB
foldseek createindex targetDB tmp #OPTIONAL generates and stores the index on disk
foldseek easy-search example/d1asha_ targetDB aln.m8 tmpFolder
The easy-cluster algorithm is designed for structural clustering by assigning structures to a representative protein structure using structural alignment. It accepts input in either PDB or mmCIF format, with support for both flat and gzipped files. By default, easy-cluster generates three output files with the following prefixes: (1) _clu.tsv, (2) _repseq.fasta, and (3) _allseq.fasta. The first file (1) is a tab-separated file describing the mapping from representative to member, while the second file (2) contains only representative sequences, and the third file (3) includes all cluster member sequences.
foldseek easy-cluster example/ res tmp -c 0.9
The provided format represents protein structure clustering in a tab-separated, two-column layout (representative and member). Each line denotes a cluster-representative and cluster-member relationship, signifying that the member shares significant structural similarity with the representative, and thus belongs to the same cluster.
Q0KJ32 Q0KJ32
Q0KJ32 C0W539
Q0KJ32 D6KVP9
E3HQM9 E3HQM9
E3HQM9 F0YHT8
The _repseq.fasta contains all representative protein sequences of the clustering.
>Q0KJ32
MAGA....R
>E3HQM9
MCAT...Q
In the _allseq.fasta file all sequences of the cluster are present. A new cluster is marked by two identical name lines of the representative sequence, where the first line stands for the cluster and the second is the name line of the first cluster sequence. It is followed by the fasta formatted sequences of all its members.
>Q0KJ32
>Q0KJ32
MAGA....R
>C0W539
MVGA....R
>D6KVP9
MVGA....R
>D1Y890
MVGV....R
>E3HQM9
>E3HQM9
MCAT...Q
>Q223C0
MCAR...Q
| Option | Category | Description |
|---|---|---|
| -e | Sensitivity | List matches below this E-value (range 0.0-inf, default: 0.001); increasing it reports more distant structures |
| --alignment-type | Alignment | 0: 3Di Gotoh-Smith-Waterman (local, not recommended), 1: TMalign (global, slow), 2: 3Di+AA Gotoh-Smith-Waterman (local, default) |
| -c | Alignment | List matches above this fraction of aligned (covered) residues (see --cov-mode) (default: 0.0); higher coverage = more global alignment |
| --cov-mode | Alignment | 0: coverage of query and target, 1: coverage of target, 2: coverage of query |
| --min-seq-id | Alignment | the minimum sequence identity to be clustered |
| --tmscore-threshold | Alignment | accept alignments with an alignment TMscore > thr |
| --tmscore-threshold-mode | Alignment | normalize TMscore by 0: alignment, 1: representative, 2: member length |
| --lddt-threshold | Alignment | accept alignments with an alignment LDDT score > thr |
The easy-multimersearch module is designed for querying one or more protein complex (multi-chain) structures (supported input formats: PDB/mmCIF, flat or gzipped) against a target database of protein complex structures. It reports the similarity metrices between the complexes (e.g., the TMscore).
The examples below use files that can be found in the example directory, which is part of the Foldseek repo, if you clone it.
If you use the precompiled version of the software, you can download the files directly: 1tim.pdb.gz and 8tim.pdb.gz.
For a pairwise alignment of complexes using easy-multimersearch, run the following command:
foldseek easy-multimersearch example/1tim.pdb.gz example/8tim.pdb.gz result tmpFolder
Foldseek easy-multimersearch can also be used for searching one or more query complexes against a target database:
foldseek databases PDB pdb tmp
foldseek easy-multimersearch example/1tim.pdb.gz pdb result tmpFolder
By default, easy-multimersearch reports the output alignment in a tab-separated file.
The default output fields are: query,target,fident,alnlen,mismatch,gapopen,qstart,qend,tstart,tend,evalue,bits,complexassignid but they can be customized with the --format-output option e.g., --format-output "query,target,complexqtmscore,complexttmscore,complexassignid" alters the output to show specific scores and identifiers.
| Code | Description |
|---|---|
| Commons | |
| query | Query sequence identifier |
| target | Target sequence identifier |
| Only for scorecomplex | |
| complexqtmscore | TM-score of Complex alignment normalized by the query length |
| complexttmscore | TM-score of Complex alignment normalized by the target length |
| complexu | Rotation matrix of Complex alignment (computed to by TM-score) |
| complext | Translation vector of Complex alignment (computed to by TM-score) |
| complexassignid | Index of Complex alignment |
Example Output:
1tim.pdb.gz_A 8tim.pdb.gz_A 0.967 247 8 0 1 247 1 247 5.412E-43 1527 0
1tim.pdb.gz_B 8tim.pdb.gz_B 0.967 247 8 0 1 247 1 247 1.050E-43 1551 0
easy-multimersearch also generates a report (prefixed _report), which provides a summary of the inter-complex chain matching, including identifiers, chains, TMscores, rotation matrices, translation vectors, and assignment IDs. The report includes the following fields:
| Column | Description |
|---|---|
| 1 | Identifier of the query complex |
| 2 | Identifier of the target complex |
| 3 | Comma separated matched chains in the query complex |
| 4 | Comma separated matched chains in the target complex |
| 5 | TM score normalized by query length [0-1] |
| 6 | TM score normalized by target length [0-1] |
| 7 | Comma separated nine rotation matrix (U) values |
| 8 | Comma separated three translation vector (T) values |
| 9 | Complex alignment ID |
Example Output:
1tim.pdb.gz 8tim.pdb.gz A,B A,B 0.98941 0.98941 0.999983,0.000332,0.005813,-0.000373,0.999976,0.006884,-0.005811,-0.006886,0.999959 0.298992,0.060047,0.565875 0
The easy-multimercluster module is designed for multimer-level structural clustering(supported input formats: PDB/mmCIF, flat or gzipped). By default, easy-multimercluster generates three output files with the following prefixes: (1) _cluster.tsv, (2) _rep_seq.fasta and (3) _cluster_report. The first file (1) is a tab-separated file describing the mapping from representative multimer to member, while the second file (2) contains only representative sequences. The third file (3) is also a tab-separated file describing filtered alignments.
Make sure chain names in PDB/mmcIF files does not contain underscores(_).
foldseek easy-multimercluster example/ clu tmp --multimer-tm-threshold 0.65 --chain-tm-threshold 0.5 --interface-lddt-threshold 0.65
5o002 5o002
194l2 194l2
194l2 193l2
10mh121 10mh121
10mh121 10mh114
10mh121 10mh119
#5o002
>5o002_A
SHGK...R
>5o002_B
SHGK...R
#194l2
>194l2_A0
KVFG...L
>194l2_A6
KVFG...L
#10mh121
...
The _cluster_report contains qcoverage, tcoverage, multimer qTm, multimer tTm, interface lddt, ustring, tstring of alignments after filtering and before clustering.
5o0f2 5o0f2 1.000 1.000 1.000 1.000 1.000 1.000,0.000,0.000,0.000,1.000,0.000,0.000,0.000,1.000 0.000,0.000,0.000
5o0f2 5o0d2 1.000 1.000 0.999 0.992 1.000 0.999,0.000,-0.000,-0.000,0.999,-0.000,0.000,0.000,0.999 -0.004,-0.001,0.084
5o0f2 5o082 1.000 0.990 0.978 0.962 0.921 0.999,-0.025,-0.002,0.025,0.999,-0.001,0.002,0.001,0.999 -0.039,0.000,-0.253
The query and target coverages here represent the sum of the coverages of all aligned chains, divided by the total query and target multimer length respectively.
| Option | Category | Description |
|---|---|---|
| -e | Sensitivity | List matches below this E-value (range 0.0-inf, default: 0.001); increasing it reports more distant structures |
| --alignment-type | Alignment | 0: 3Di Gotoh-Smith-Waterman (local, not recommended), 1: TMalign (global, slow), 2: 3Di+AA Gotoh-Smith-Waterman (local, default) |
| -c | Alignment | List matches above this fraction of aligned (covered) residues (see --cov-mode) (default: 0.0); higher coverage = more global alignment |
| --cov-mode | Alignment | 0: coverage of query and target (cluster multimers only with same chain numbers), 1: coverage of target, 2: coverage of query |
| --multimer-tm-threshold | Alignment | accept alignments with multimer alignment TMscore > thr |
| --chain-tm-threshold | Alignment | accept alignments if every single chain TMscore > thr |
| --interface-lddt-threshold | Alignment | accept alignments with an interface LDDT score > thr |
easy-searchfast protein structure searcheasy-clusterfast protein structure clusteringeasy-multimersearchfast protein multimer-level structure searcheasy-multimerclusterfast protein multimer-level structure clusteringcreatedbcreate a database from protein structures (PDB,mmCIF, mmJSON)databasesdownload pre-assembled databases
The easiest way to get the alignment TMscore normalized by min(alnLen,qLen,targetLen) as well as a rotation matrix is through the following command:
foldseek easy-search example/ example/ aln tmp --format-output query,target,alntmscore,u,t
Alternatively, it is possible to compute TMscores for the kind of alignment output (e.g., 3Di+AA) using the following commands:
foldseek createdb example/ targetDB
foldseek createdb example/ queryDB
foldseek search queryDB targetDB aln tmpFolder -a
foldseek aln2tmscore queryDB targetDB aln aln_tmscore
foldseek createtsv queryDB targetDB aln_tmscore aln_tmscore.tsv
Output format aln_tmscore.tsv: query and target identifiers, TMscore, translation(3) and rotation vector=(3x3)
Foldseek can output multiple sequence alignments in a3m format using the following commands.
To convert a3m to FASTA format, the following script can be used reformat.pl (reformat.pl in.a3m out.fas).
foldseek createdb example/ targetDB
foldseek createdb example/ queryDB
foldseek search queryDB targetDB aln tmpFolder -a
foldseek result2msa queryDB targetDB aln msa --msa-format-mode 6
foldseek unpackdb msa msa_output --unpack-suffix a3m --unpack-name-mode 0